Note
This tutorial was generated from an IPython notebook that can be downloaded here.
# ref: https://radvel.readthedocs.io/en/latest/tutorials/K2-24_Fitting+MCMC.html
# ref: https://arxiv.org/abs/1511.04497
%matplotlib inline
%config InlineBackend.figure_format = "retina"
from matplotlib import rcParams
rcParams["savefig.dpi"] = 100
rcParams["figure.dpi"] = 100
rcParams["font.size"] = 16
rcParams["text.usetex"] = False
rcParams["font.family"] = ["sans-serif"]
rcParams["font.sans-serif"] = ["cmss10"]
rcParams["axes.unicode_minus"] = False
import corner
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from scipy.signal import savgol_filter
from astropy.io import fits
from astropy.stats import BoxLeastSquares, LombScargle
from astropy import constants, units
import pymc3 as pm
import pymc3.distributions.transforms as tr
import theano
import theano.tensor as tt
import exoplanet
from exoplanet import distributions
from exoplanet.gp import terms, GP
lc_url = "https://archive.stsci.edu/hlsps/everest/v2/c02/203700000/71098/hlsp_everest_k2_llc_203771098-c02_kepler_v2.0_lc.fits"
with fits.open(lc_url) as hdus:
lc = hdus[1].data
lc_hdr = hdus[1].header
texp = lc_hdr["FRAMETIM"] * lc_hdr["NUM_FRM"]
texp /= 60.0 * 60.0 * 24.0
m = (np.arange(len(lc)) > 100) & np.isfinite(lc["FLUX"]) & np.isfinite(lc["TIME"])
bad_bits=[1, 2, 3, 4, 5, 6, 7, 8, 9, 11, 12, 13, 14, 16, 17]
qual = lc["QUALITY"]
for b in bad_bits:
m &= qual & 2 ** (b - 1) == 0
x = lc["TIME"][m]
y = lc["FLUX"][m]
mu = np.median(y)
y = (y / mu - 1) * 1e3
smooth = savgol_filter(y, 501, polyorder=5)
resid = y - smooth
sigma = np.sqrt(np.mean(resid**2))
m = resid < 3 * sigma
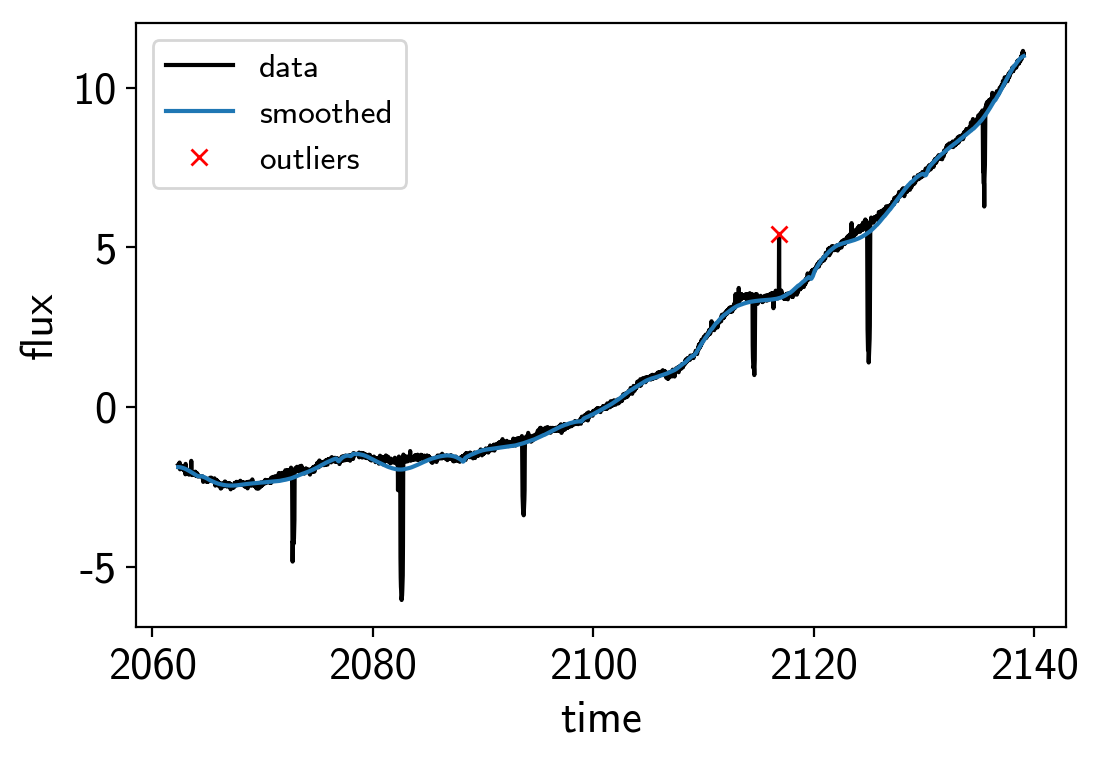
plt.plot(x, y, "k", label="data")
plt.plot(x, smooth, label="smoothed")
plt.plot(x[~m], y[~m], "xr", label="outliers")
plt.legend(fontsize=12)
plt.xlabel("time")
plt.ylabel("flux")
x_ref = np.min(x[m])
x = np.ascontiguousarray(x[m], dtype=np.float64)
x -= x_ref
y = np.ascontiguousarray(y[m], dtype=np.float64)
smooth = savgol_filter(y, 501, polyorder=5)
data = pd.read_csv("https://raw.githubusercontent.com/California-Planet-Search/radvel/master/example_data/epic203771098.csv", index_col=0)
rv_x = np.array(data.t - x_ref)
rv_y = np.array(data.vel)
rv_yerr = np.array(data.errvel)
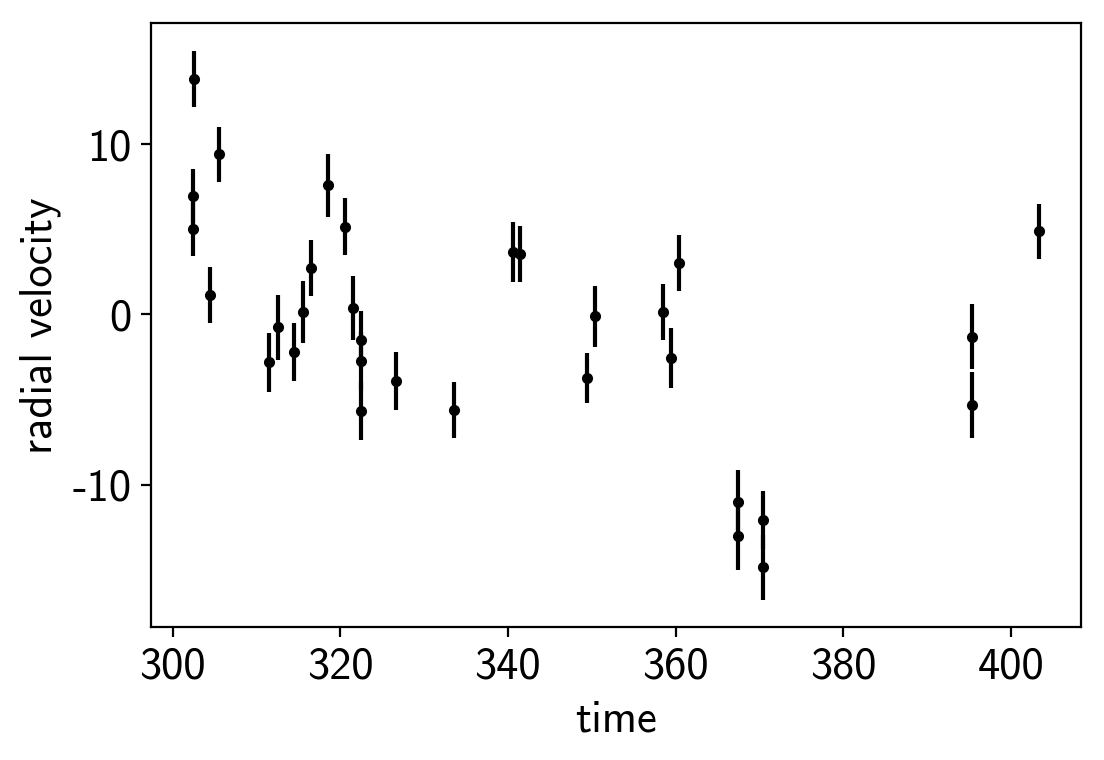
plt.errorbar(rv_x, rv_y, yerr=rv_yerr, fmt=".k")
plt.xlabel("time")
plt.ylabel("radial velocity");
m = np.zeros(len(x), dtype=bool)
period_grid = np.exp(np.linspace(np.log(10), np.log(50), 50000))
bls_results = []
periods = []
t0s = []
depths = []
for i in range(2):
bls = BoxLeastSquares(x[~m], y[~m] - smooth[~m])
bls_power = bls.power(period_grid, 0.1, oversample=20)
bls_results.append(bls_power)
index = np.argmax(bls_power.power)
periods.append(bls_power.period[index])
t0s.append(bls_power.transit_time[index])
depths.append(bls_power.depth[index])
m |= bls.transit_mask(x, periods[-1], 0.5, t0s[-1])
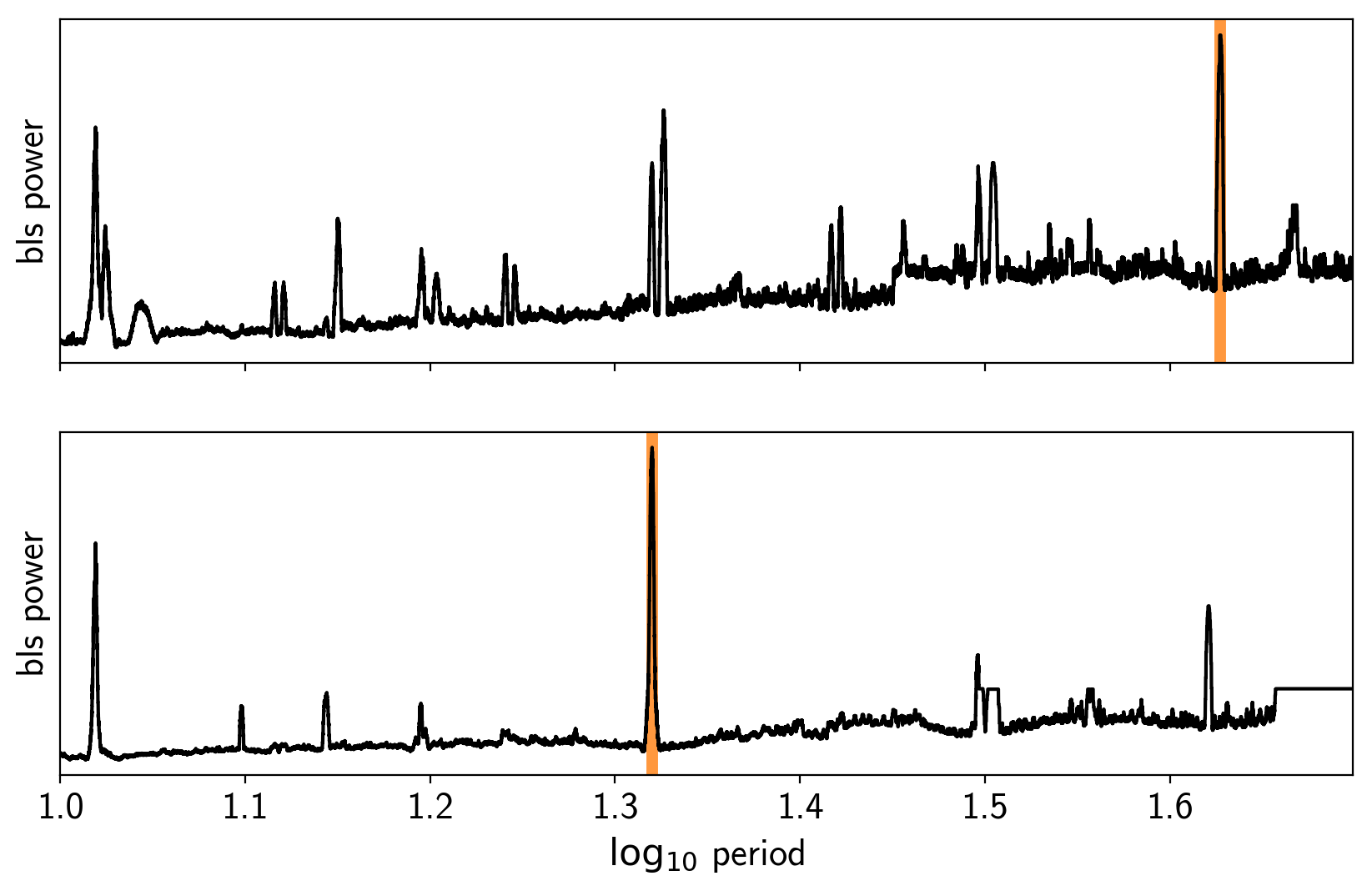
fig, axes = plt.subplots(len(bls_results), 1, figsize=(10, 6), sharex=True)
for i in range(len(bls_results)):
ax = axes[i]
ax.axvline(np.log10(periods[i]), color="C1", lw=5, alpha=0.8)
ax.plot(np.log10(bls_results[i].period), bls_results[i].power, "k")
ax.set_ylabel("bls power")
ax.set_yticks([])
ax = axes[-1]
ax.set_xlim(np.log10(period_grid.min()), np.log10(period_grid.max()))
ax.set_xlabel("$\log_{10}$ period");
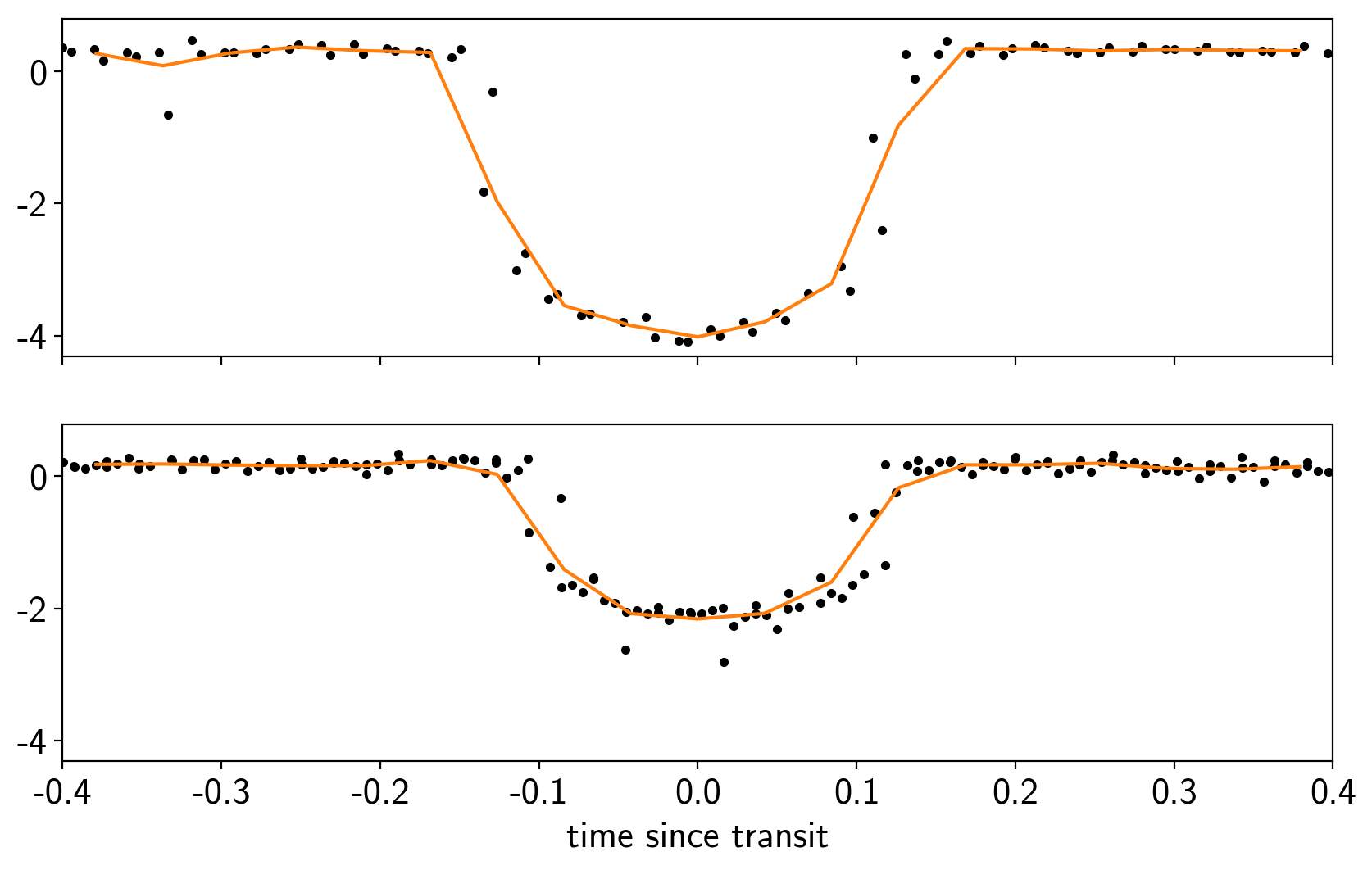
fig, axes = plt.subplots(len(bls_results), 1, figsize=(10, 6), sharex=True)
for i in range(len(bls_results)):
ax = axes[i]
p = periods[i]
x_fold = (x - t0s[i] + 0.5*p) % p - 0.5*p
ax.plot(x_fold, y - smooth, ".k")
bins = np.linspace(-0.4, 0.4, 20)
denom, _ = np.histogram(x_fold, bins)
num, _ = np.histogram(x_fold, bins, weights=y - smooth)
denom[num == 0] = 1.0
ax.plot(0.5*(bins[1:] + bins[:-1]), num / denom, color="C1")
ax = axes[-1]
ax.set_xlim(-0.4, 0.4)
ax.set_xlabel("time since transit");
msini = exoplanet.estimators.estimate_minimum_mass(periods, rv_x, rv_y, rv_yerr, t0s=t0s).to(units.M_earth)
print(msini)
[30.3178443 22.08252394] earthMass
N_planets = len(periods)
rv_x_grid = np.linspace(rv_x.min(), rv_x.max(), 1000)
def build_model(mask=None):
if mask is None:
mask = np.ones(len(x), dtype=bool)
with pm.Model() as model:
# Stellar properties
mean = pm.Normal("mean", mu=0.0, sd=1.0)
u = distributions.Triangle("u", shape=2)
mstar = pm.Bound(pm.Normal, lower=0.0)("mstar", mu=1.12, sd=0.05)
rstar = pm.Bound(pm.Normal, lower=0.0)("rstar", mu=1.21, sd=0.11)
t0 = pm.Normal("t0", mu=t0s, sd=0.1, shape=N_planets)
logperiod = pm.Normal("logperiod", mu=np.log(periods), sd=1e-4, shape=N_planets)
period = pm.Deterministic("period", tt.exp(logperiod))
logm = pm.Uniform(
"logm",
lower=np.log(10.0),
upper=np.log(50.0),
shape=N_planets, testval=np.log(msini.value))
m = pm.Deterministic("m", tt.exp(logm))
# Radius/impact parameter joint distribution
rb_test = 0.5 + np.zeros((2, N_planets))
rb_test[0, :] = np.sqrt(1e-3)*np.sqrt(depths)
rb = distributions.RadiusImpactParameter(
"rb", min_radius=0.01, max_radius=0.1,
shape=(2, N_planets), testval=rb_test)
ror = pm.Deterministic("ror", rb[0])
r = pm.Deterministic("r", ror * rstar)
b = pm.Deterministic("b", rb[1])
# Log-uniform prior on r
pm.Potential("logrprior", -tt.log(r))
ecc = pm.Beta("ecc", alpha=0.867, beta=3.03, shape=N_planets,
testval=np.array([0.1, 0.1]))
omega = distributions.Angle("omega", shape=N_planets)
logjitter2 = pm.Normal("logjitter2", mu=np.log(0.05), sd=5.0)
v0 = pm.Normal("v0", mu=0.0, sd=1.0)
dvdt = pm.Normal("dvdt", mu=0.0, sd=0.1)
d2vdt2 = pm.Normal("d2vdt2", mu=0.0, sd=0.01)
# Set up the orbit
orbit = exoplanet.orbits.KeplerianOrbit(
period=period,
t0=t0,
b=b,
ecc=ecc,
omega=omega,
m_planet=m,
m_planet_units=msini.unit,
r_star=rstar,
m_star=mstar,
)
pm.Deterministic("incl", orbit.incl)
pm.Deterministic("a", orbit.a)
lc = exoplanet.StarryLightCurve(u, r_star=rstar)
transit = lc.get_light_curve(r, orbit, x[mask], texp=texp, oversample=5, order=2) * 1e3
# transit = tt.zeros(len(y))
# transit = tt.set_subtensor(transit[mask_near], tt.sum(transits, axis=-1) * 1e3)
pm.Deterministic("transit", transit)
# Likelihood
logs2 = pm.Normal("logs2", mu=-5.0, sd=5.0)
logw0 = pm.Normal("logw0", mu=0.0, sd=5.0)
logQ = pm.Normal("logQ", mu=1.0, sd=5.0)
delta = pm.Normal("delta", mu=0.0, sd=10.0)
logS0 = pm.Deterministic("logS0", delta - logw0 - logQ)
kernel = terms.SHOTerm(S0=tt.exp(logS0), w0=tt.exp(logw0), Q=tt.exp(logQ))
gp = GP(kernel, x[mask], tt.exp(logs2) + np.zeros_like(x[mask]), J=2)
resid = y[mask] - transit - mean
pm.Potential("obs", gp.log_likelihood(resid))
gp_pred = gp.predict()
pm.Deterministic("gp", mean + gp_pred)
pm.Deterministic("resid", resid - gp_pred)
# RV model
# Convert R_sun / day to m / s
rv_x_mid = 0.5 * (rv_x.max() + rv_x.min())
rv_dx = rv_x - rv_x_mid
rv_dx_grid = rv_x_grid - rv_x_mid
rv_conv = (1 * units.R_sun / units.day).to(units.m / units.s).value
vrad = rv_conv * tt.sum(orbit.get_star_velocity(rv_x)[2], axis=-1)
bkg = v0 + dvdt * rv_dx + d2vdt2 * rv_dx**2
pm.Normal("rv_obs", mu=vrad + bkg, sd=tt.sqrt(rv_yerr**2 + tt.exp(logjitter2)), observed=rv_y)
pm.Deterministic("vrad", rv_conv * orbit.get_star_velocity(rv_x_grid)[2])
pm.Deterministic("bkg", v0 + dvdt * rv_dx_grid + d2vdt2 * rv_dx_grid**2)
soln = pm.find_MAP(start=model.test_point, vars=[logs2, delta, logw0, logQ, mean])
soln = pm.find_MAP(start=soln, vars=[logperiod, t0])
soln = pm.find_MAP(start=soln)
return model, soln
model0, soln0 = build_model()
logp = 1,875.4, ||grad|| = 1.5226: 100%|██████████| 27/27 [00:00<00:00, 211.07it/s]
logp = 2,069.1, ||grad|| = 702.99: 100%|██████████| 33/33 [00:00<00:00, 58.22it/s]
logp = 4,711.9, ||grad|| = 1,894.7: 100%|██████████| 3307/3307 [00:22<00:00, 143.83it/s]
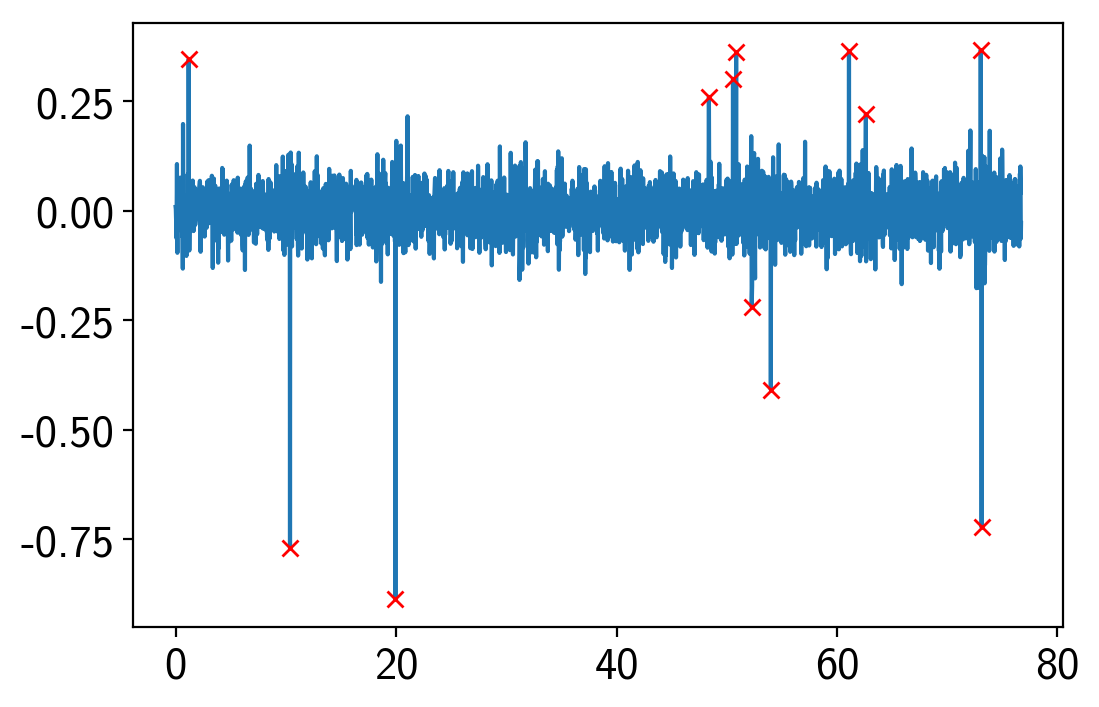
# Sigma clipping
resid = soln0["resid"]
sigma = np.sqrt(np.mean(resid**2))
mask = np.abs(resid) < 4 * sigma
plt.plot(x, resid)
plt.plot(x[~mask], resid[~mask], "xr")
[<matplotlib.lines.Line2D at 0x1c3b66af98>]
model, soln = build_model(mask)
logp = 1,929.7, ||grad|| = 0.0020057: 100%|██████████| 33/33 [00:00<00:00, 245.05it/s]
logp = 2,143, ||grad|| = 2,207.3: 100%|██████████| 45/45 [00:00<00:00, 93.59it/s]
logp = 5,246.1, ||grad|| = 776.46: 100%|██████████| 3354/3354 [00:20<00:00, 164.11it/s]
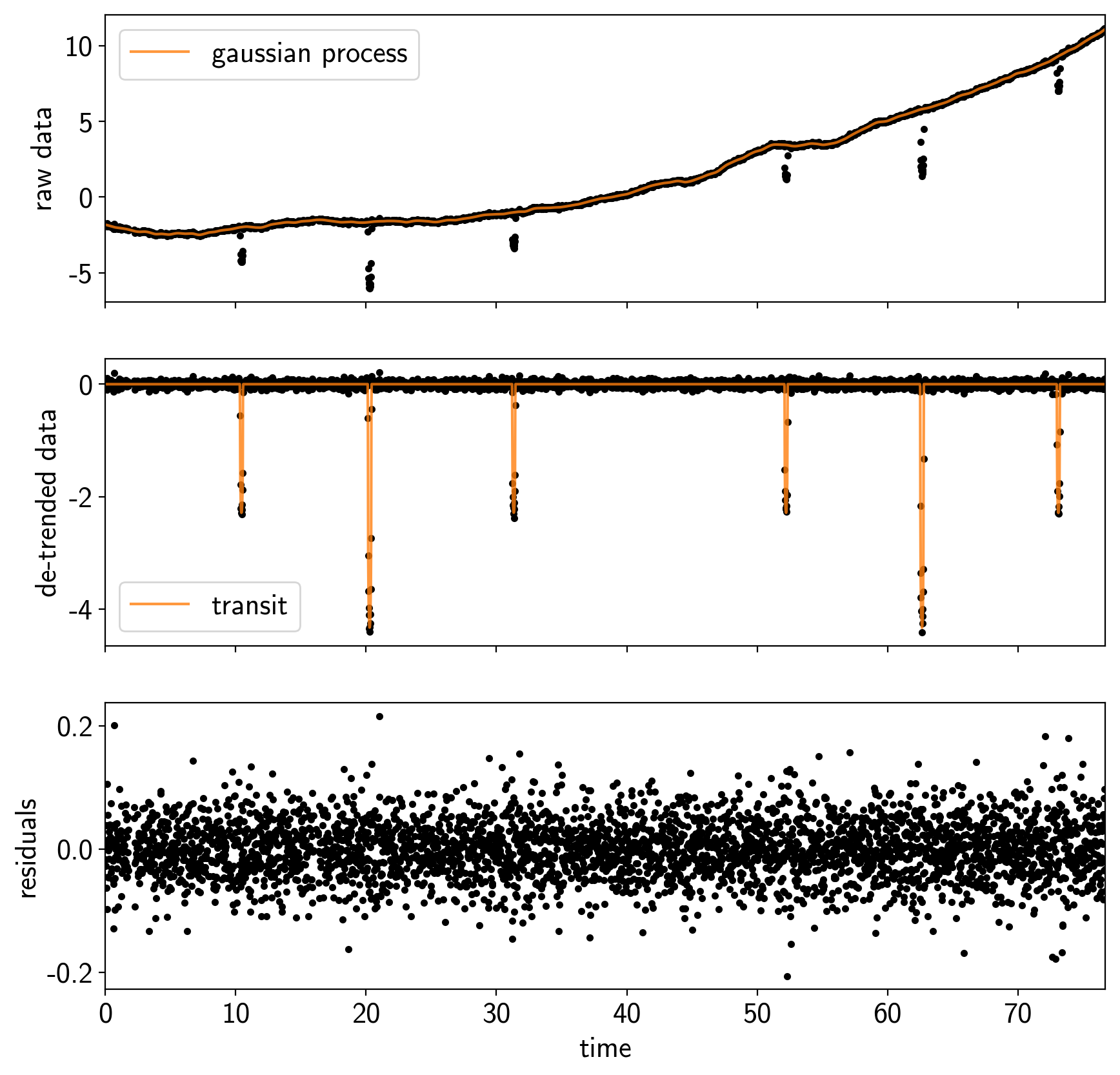
fig, axes = plt.subplots(3, 1, figsize=(10, 10), sharex=True)
ax = axes[0]
ax.plot(x[mask], y[mask], ".k")
ax.plot(x[mask], soln["gp"], color="C1", alpha=0.8, label="gaussian process")
ax.set_ylabel("raw data")
ax.legend()
ax = axes[1]
ax.plot(x[mask], y[mask] - soln["gp"], ".k")
ax.plot(x[mask], soln["transit"], color="C1", alpha=0.8, label="transit")
ax.set_ylabel("de-trended data")
ax.legend()
ax = axes[2]
ax.plot(x[mask], soln["resid"], ".k")
ax.set_ylabel("residuals")
ax.set_xlabel("time")
ax.set_xlim(x[mask].min(), x[mask].max());
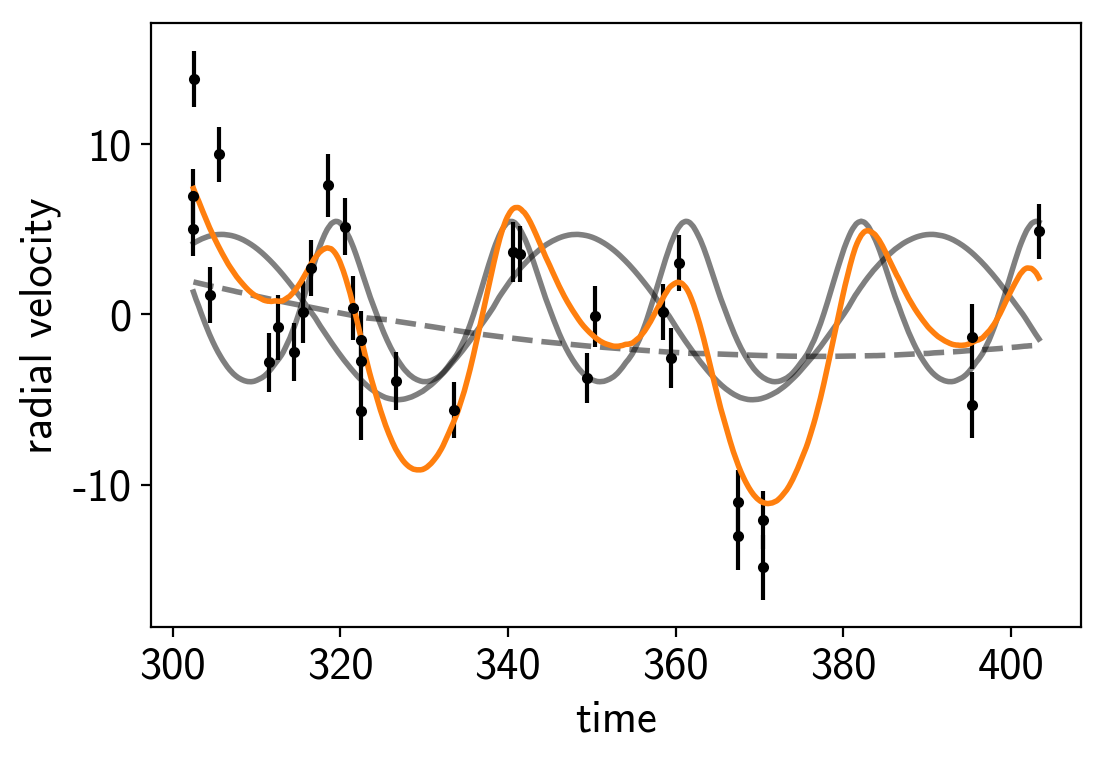
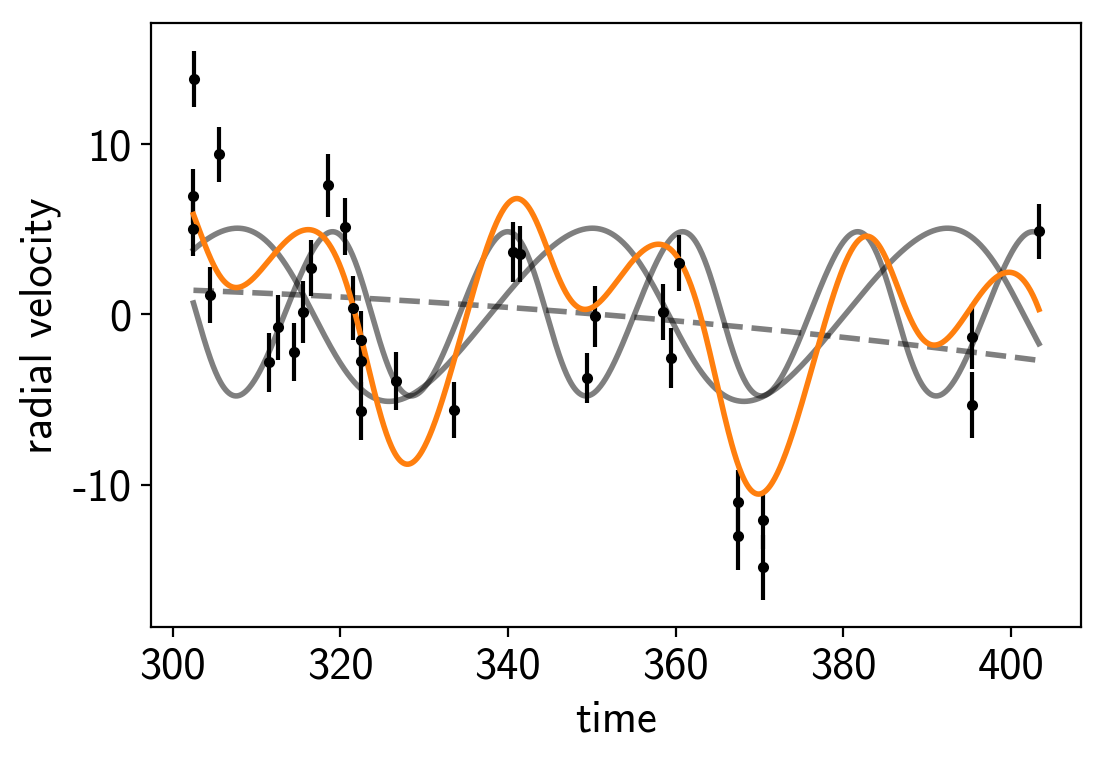
plt.plot(rv_x_grid, soln["vrad"], color="k", lw=2, alpha=0.5)
plt.plot(rv_x_grid, soln["bkg"], "--", color="k", lw=2, alpha=0.5)
plt.plot(rv_x_grid, np.sum(soln["vrad"], axis=-1) + soln["bkg"], color="C1", lw=2)
plt.errorbar(rv_x, rv_y, yerr=rv_yerr, fmt=".k")
plt.xlabel("time")
plt.ylabel("radial velocity")
Text(0,0.5,'radial velocity')
schedule = exoplanet.sampling.TuningSchedule(start=50, window=50)
with model:
burnin = schedule.tune(tune=2000, start=soln, step_kwargs=dict(regular_window=0))
Only 2 samples in chain.
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [delta, logQ, logw0, logs2, d2vdt2, dvdt, v0, logjitter2, omega, ecc, rb, logm, logperiod, t0, rstar, mstar, u, mean]
Sampling 2 chains: 100%|██████████| 104/104 [00:44<00:00, 1.05s/draws]
The chain reached the maximum tree depth. Increase max_treedepth, increase target_accept or reparameterize.
The chain reached the maximum tree depth. Increase max_treedepth, increase target_accept or reparameterize.
Only 2 samples in chain.
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [delta, logQ, logw0, logs2, d2vdt2, dvdt, v0, logjitter2, omega, ecc, rb, logm, logperiod, t0, rstar, mstar, u, mean]
Sampling 2 chains: 100%|██████████| 104/104 [00:49<00:00, 1.03s/draws]
The chain reached the maximum tree depth. Increase max_treedepth, increase target_accept or reparameterize.
The chain reached the maximum tree depth. Increase max_treedepth, increase target_accept or reparameterize.
Only 2 samples in chain.
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [delta, logQ, logw0, logs2, d2vdt2, dvdt, v0, logjitter2, omega, ecc, rb, logm, logperiod, t0, rstar, mstar, u, mean]
Sampling 2 chains: 100%|██████████| 204/204 [01:42<00:00, 1.79s/draws]
The chain reached the maximum tree depth. Increase max_treedepth, increase target_accept or reparameterize.
Only 2 samples in chain.
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [delta, logQ, logw0, logs2, d2vdt2, dvdt, v0, logjitter2, omega, ecc, rb, logm, logperiod, t0, rstar, mstar, u, mean]
Sampling 2 chains: 100%|██████████| 404/404 [03:37<00:00, 1.95s/draws]
Only 2 samples in chain.
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [delta, logQ, logw0, logs2, d2vdt2, dvdt, v0, logjitter2, omega, ecc, rb, logm, logperiod, t0, rstar, mstar, u, mean]
Sampling 2 chains: 100%|██████████| 804/804 [06:50<00:00, 1.28draws/s]
Only 2 samples in chain.
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [delta, logQ, logw0, logs2, d2vdt2, dvdt, v0, logjitter2, omega, ecc, rb, logm, logperiod, t0, rstar, mstar, u, mean]
Sampling 2 chains: 100%|██████████| 2404/2404 [24:13<00:00, 1.19draws/s]
plt.plot(burnin["ecc"])
[<matplotlib.lines.Line2D at 0x1c4e950518>,
<matplotlib.lines.Line2D at 0x1c4e950668>]
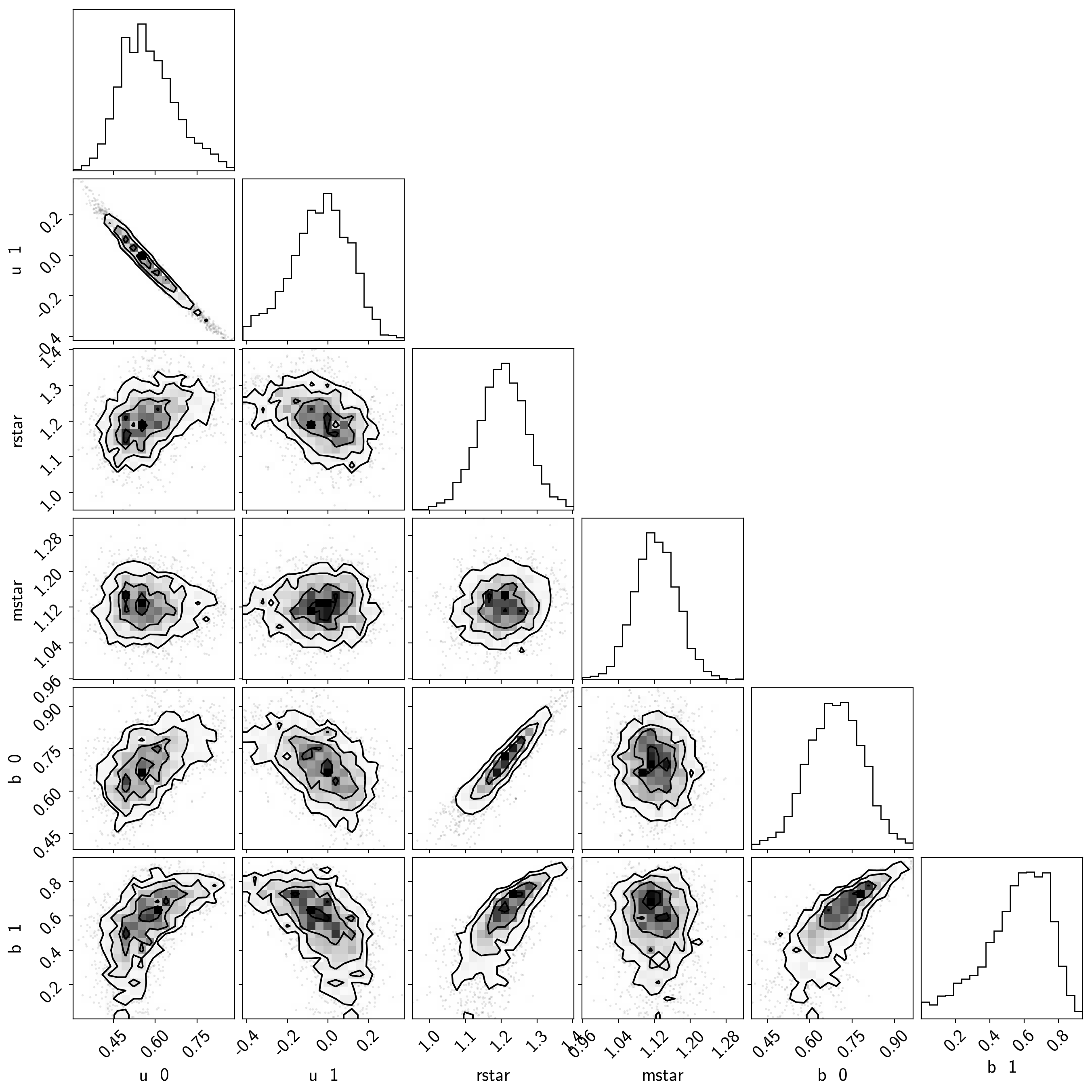
samples = pm.trace_to_dataframe(burnin, varnames=["u", "rstar", "mstar", "b"])
samples.columns = [k.replace("_", " ") for k in samples.columns]
corner.corner(samples);
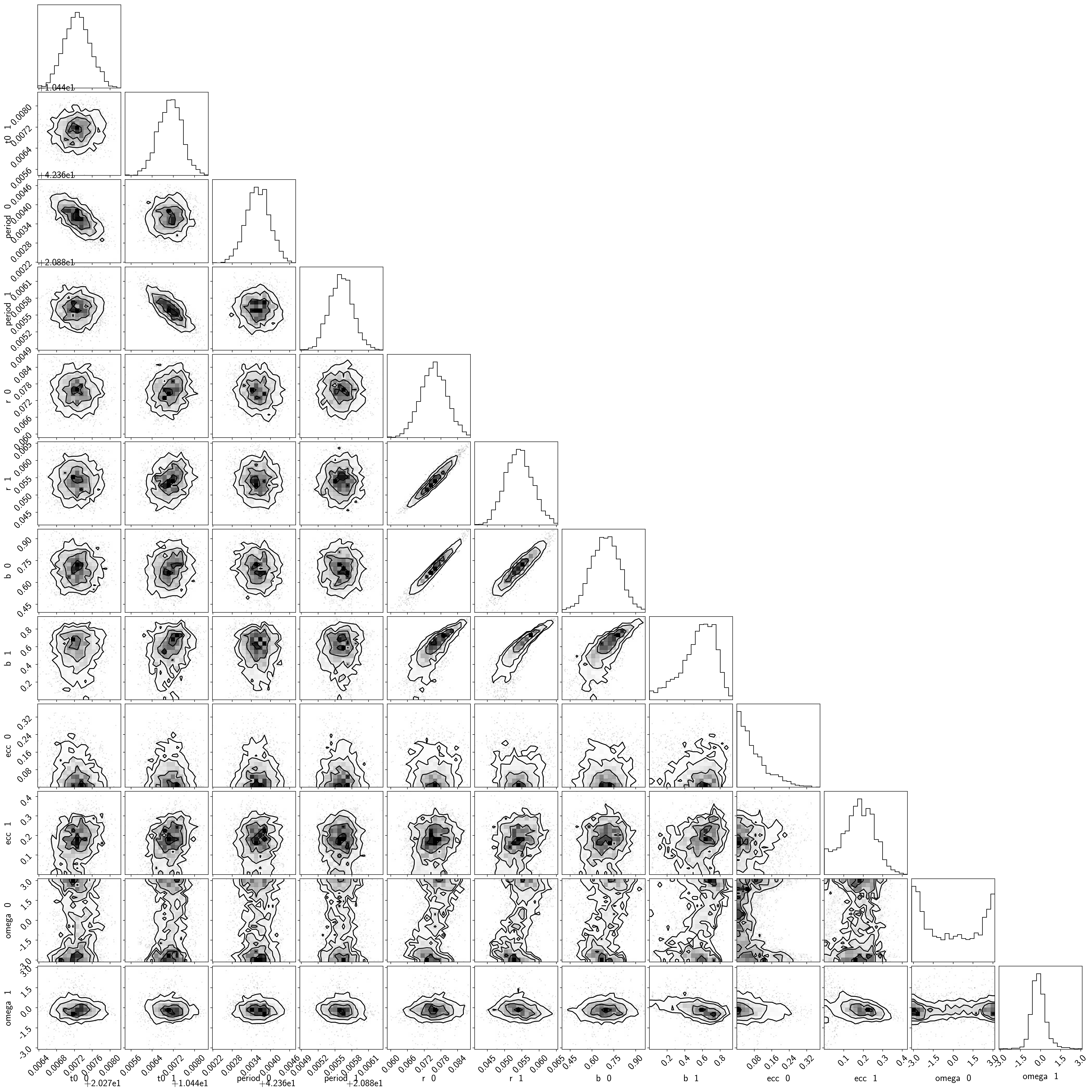
samples = pm.trace_to_dataframe(burnin, varnames=["t0", "period", "r", "b", "ecc", "omega"])
samples.columns = [k.replace("_", " ") for k in samples.columns]
corner.corner(samples);
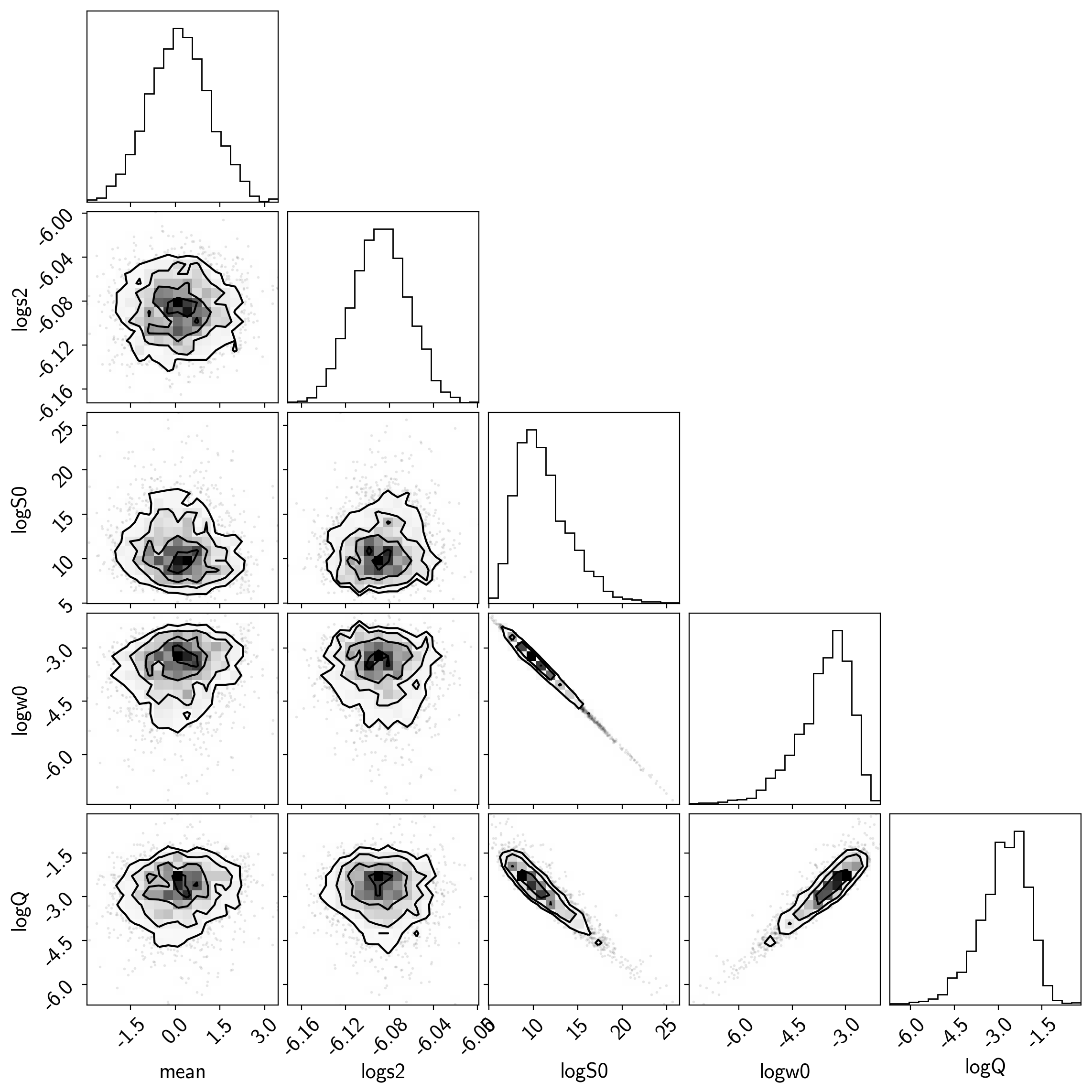
samples = pm.trace_to_dataframe(burnin, varnames=["mean", "logs2", "logS0", "logw0", "logQ"])
samples.columns = [k.replace("_", " ") for k in samples.columns]
corner.corner(samples);
with model:
trace = schedule.sample(draws=2000)
Multiprocess sampling (2 chains in 2 jobs) NUTS: [delta, logQ, logw0, logs2, d2vdt2, dvdt, v0, logjitter2, omega, ecc, rb, logm, logperiod, t0, rstar, mstar, u, mean] Sampling 2 chains: 100%|██████████| 4100/4100 [38:10<00:00, 1.23s/draws] There were 70 divergences after tuning. Increase target_accept or reparameterize. The acceptance probability does not match the target. It is 0.8850118570461237, but should be close to 0.8. Try to increase the number of tuning steps. There were 318 divergences after tuning. Increase target_accept or reparameterize. The estimated number of effective samples is smaller than 200 for some parameters.
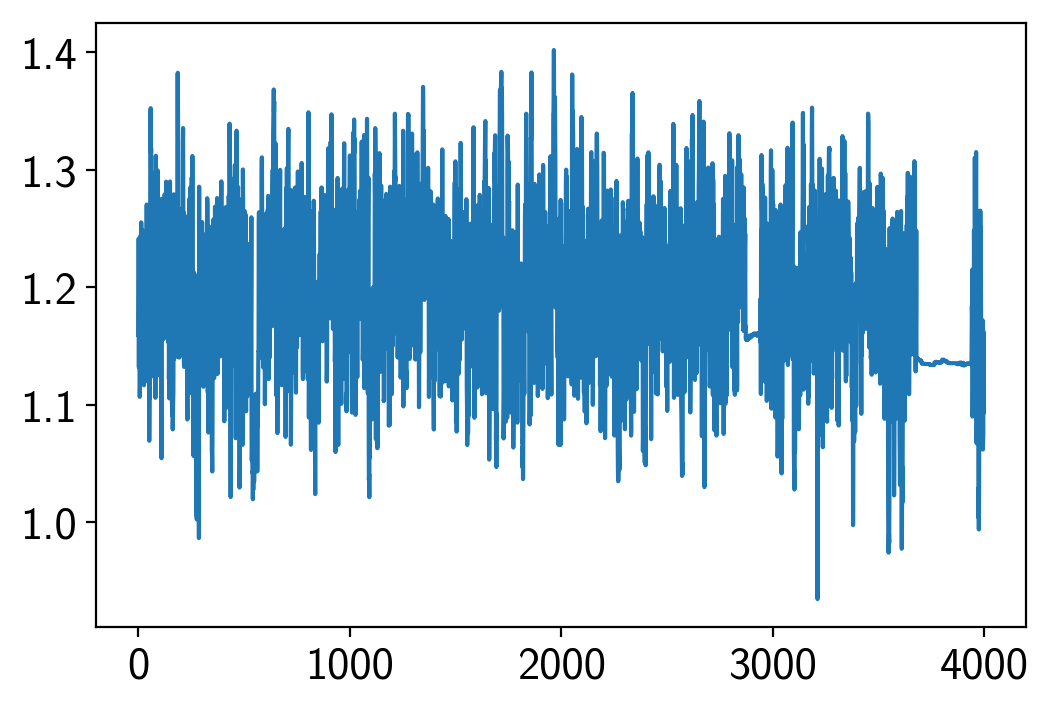
plt.plot(trace["rstar"])
[<matplotlib.lines.Line2D at 0x1c39a679b0>]
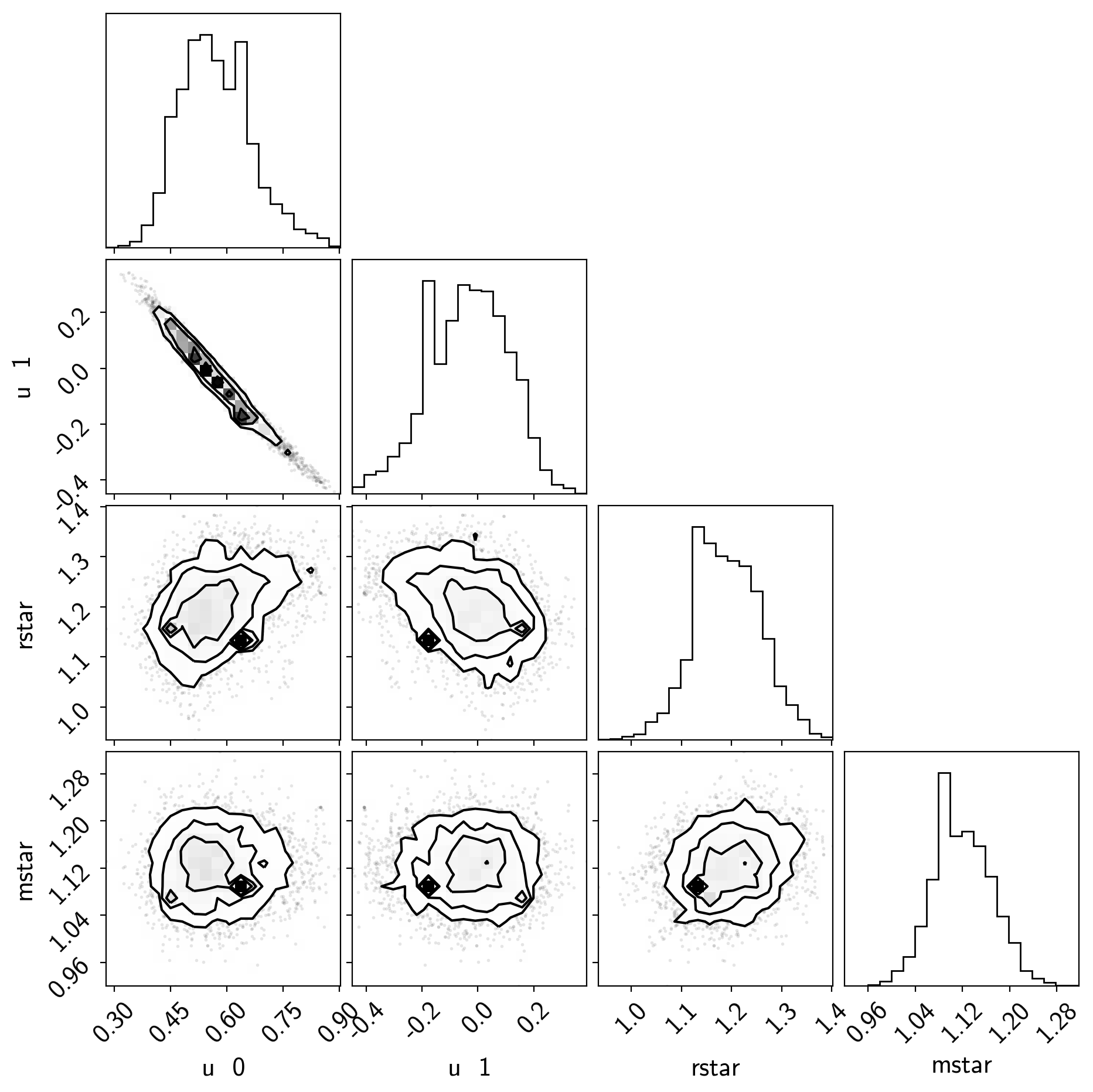
samples = pm.trace_to_dataframe(trace, varnames=["u", "rstar", "mstar"])
samples.columns = [k.replace("_", " ") for k in samples.columns]
corner.corner(samples);
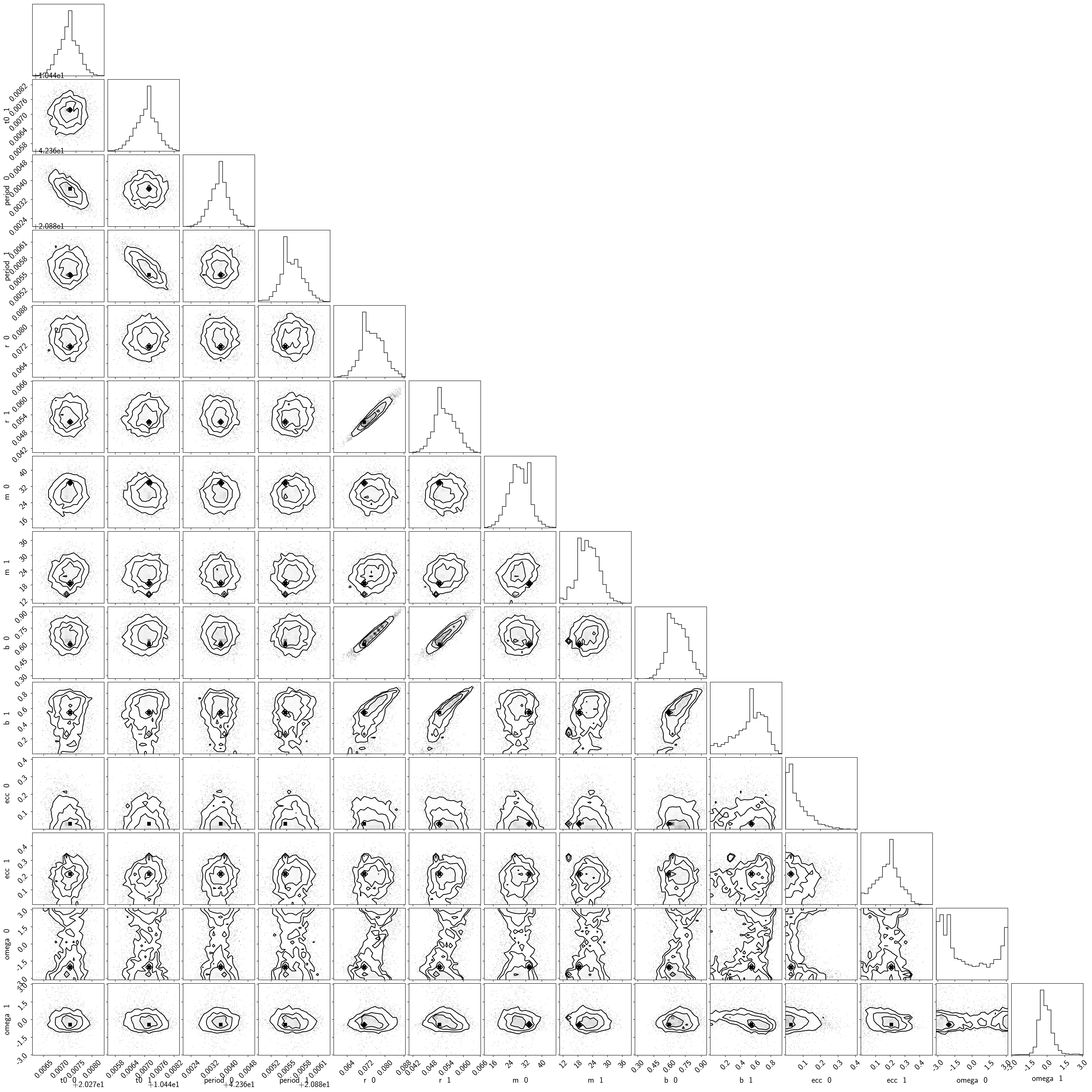
samples = pm.trace_to_dataframe(trace, varnames=["t0", "period", "r", "m", "b", "ecc", "omega"])
samples.columns = [k.replace("_", " ") for k in samples.columns]
corner.corner(samples);
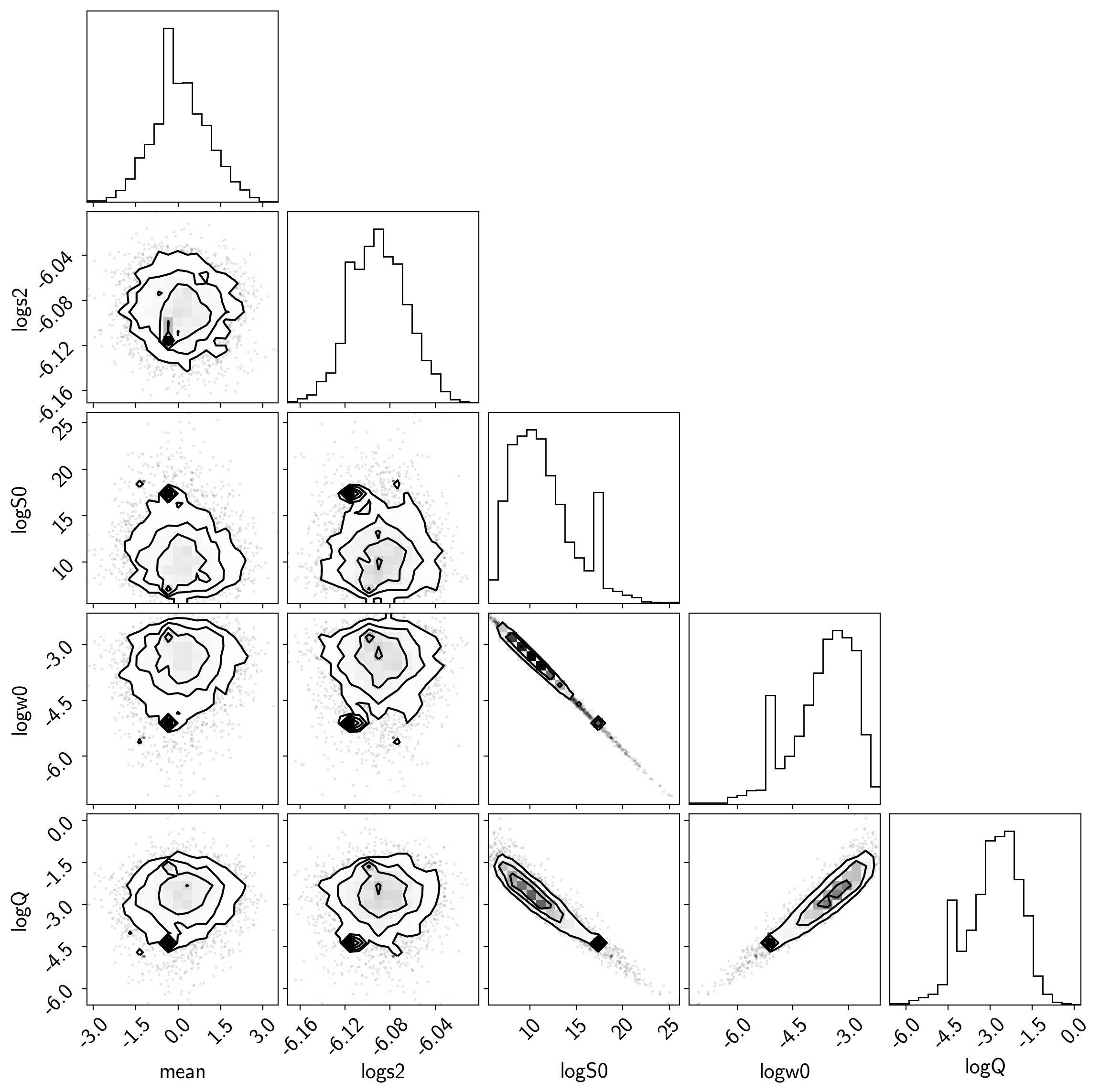
samples = pm.trace_to_dataframe(trace, varnames=["mean", "logs2", "logS0", "logw0", "logQ"])
samples.columns = [k.replace("_", " ") for k in samples.columns]
corner.corner(samples);
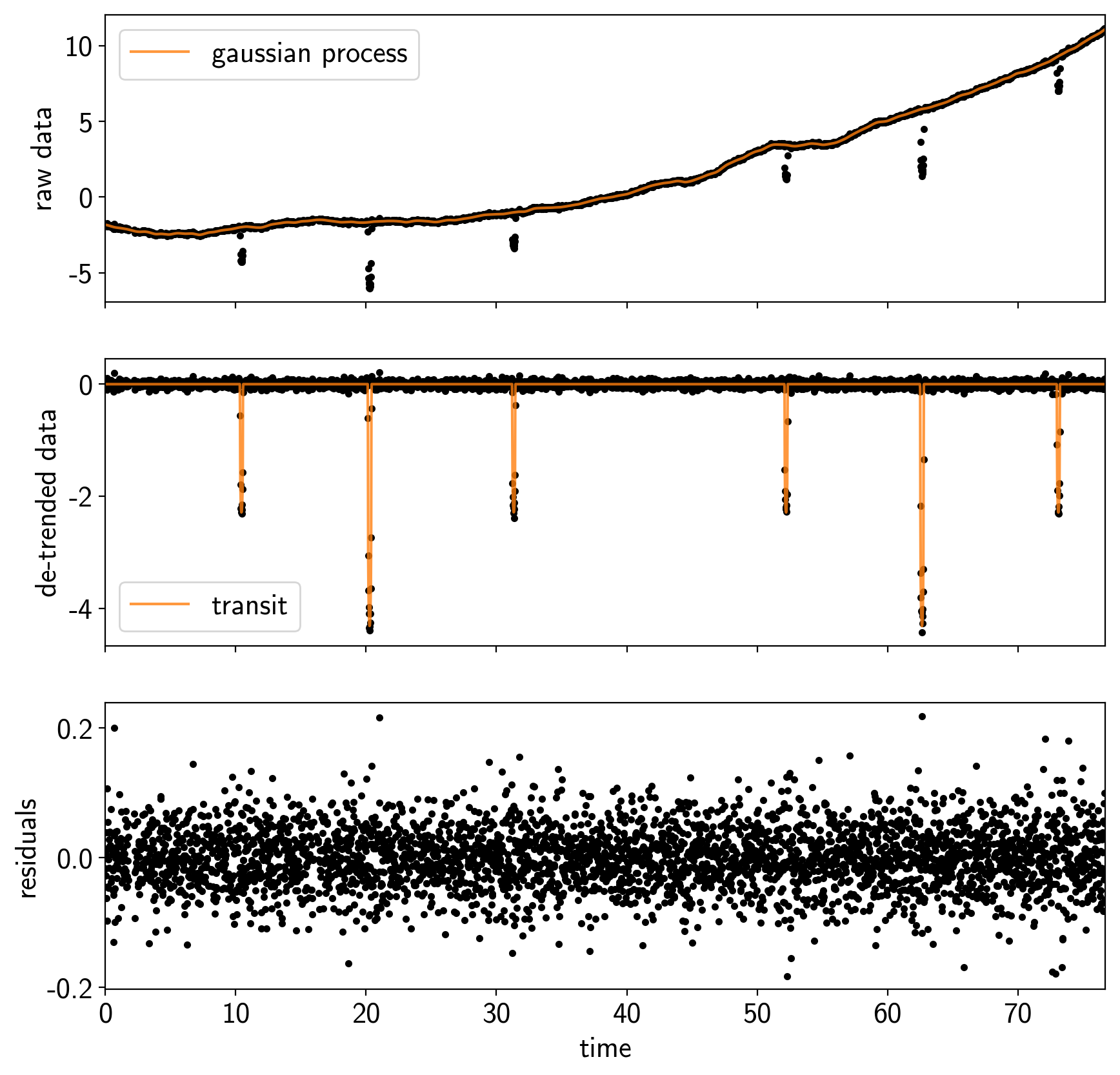
fig, axes = plt.subplots(3, 1, figsize=(10, 10), sharex=True)
gp_mod = np.median(trace["gp"], axis=0)
transit_mod = np.median(trace["transit"], axis=0)
ax = axes[0]
ax.plot(x[mask], y[mask], ".k")
ax.plot(x[mask], gp_mod, color="C1", alpha=0.8, label="gaussian process")
ax.set_ylabel("raw data")
ax.legend()
ax = axes[1]
ax.plot(x[mask], y[mask] - gp_mod, ".k")
ax.plot(x[mask], transit_mod, color="C1", alpha=0.8, label="transit")
ax.set_ylabel("de-trended data")
ax.legend()
ax = axes[2]
ax.plot(x[mask], np.median(trace["resid"], axis=0), ".k")
ax.set_ylabel("residuals")
ax.set_xlabel("time")
ax.set_xlim(x[mask].min(), x[mask].max());
vrad_mod = np.median(trace["vrad"], axis=0)
bkg_mod = np.median(trace["bkg"], axis=0)
plt.plot(rv_x_grid, vrad_mod, color="k", lw=2, alpha=0.5)
plt.plot(rv_x_grid, bkg_mod, "--", color="k", lw=2, alpha=0.5)
plt.plot(rv_x_grid, np.sum(vrad_mod, axis=-1) + bkg_mod, color="C1", lw=2)
plt.errorbar(rv_x, rv_y, yerr=rv_yerr, fmt=".k")
plt.xlabel("time")
plt.ylabel("radial velocity")
Text(0,0.5,'radial velocity')