Note
This tutorial was generated from an IPython notebook that can be downloaded here.
Transit fitting¶
exoplanet includes methods for computing the light curves transiting planets. In its simplest form this can be used to evaluate a light curve like you would do with batman, for example:
import numpy as np
import matplotlib.pyplot as plt
import exoplanet as xo
# The light curve calculation requires an orbit
orbit = xo.orbits.KeplerianOrbit(period=3.456)
# Compute a limb-darkened light curve using starry
t = np.linspace(-0.1, 0.1, 1000)
u = [0.3, 0.2]
light_curve = xo.StarryLightCurve(u).get_light_curve(
orbit=orbit, r=0.1, t=t, texp=0.02).eval()
# Note: the `eval` is needed because this is using Theano in
# the background
plt.plot(t, light_curve, color="C0", lw=2)
plt.ylabel("relative flux")
plt.xlabel("time [days]")
plt.xlim(t.min(), t.max());
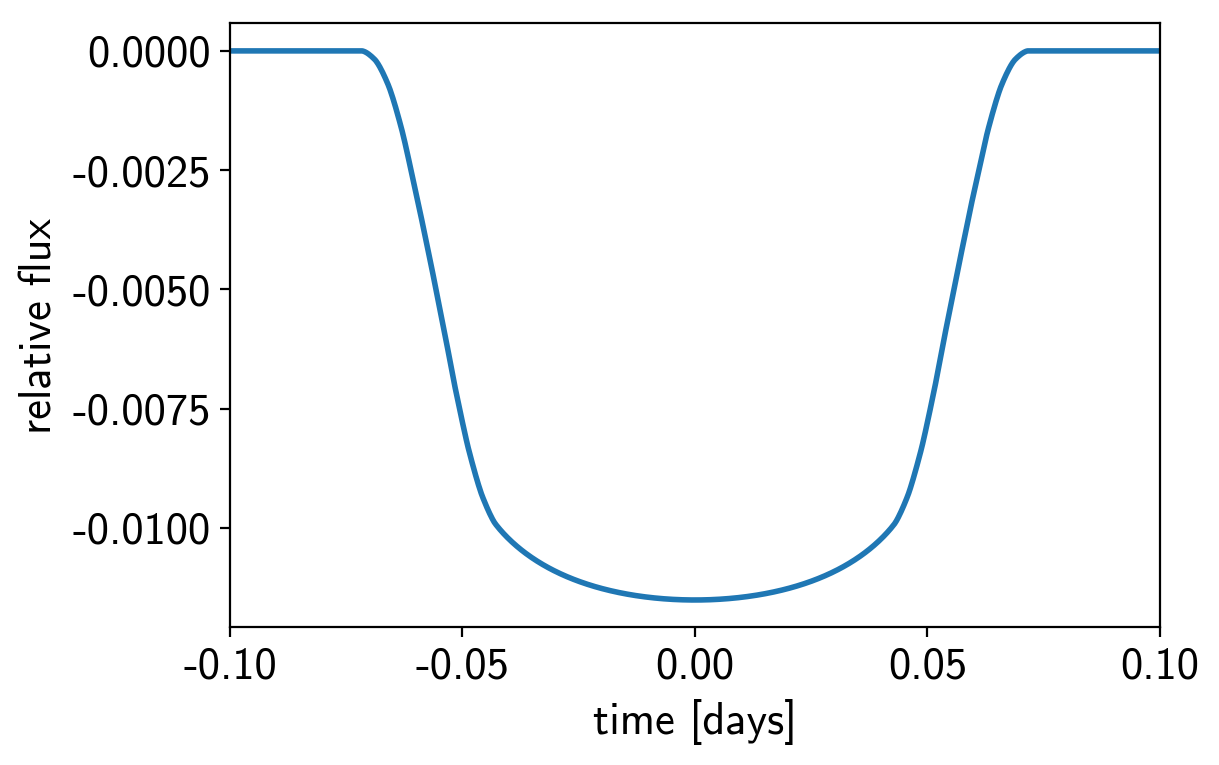
But the real power comes from the fact that this is defined as a Theano operation so it can be combined with PyMC3 to do transit inference using Hamiltonian Monte Carlo.
The transit model in PyMC3¶
In this section, we will construct a simple transit fit model using PyMC3 and then we will fit a two planet model to simulated data. To start, let’s randomly sample some periods and phases and then define the time sampling:
np.random.seed(123)
periods = np.random.uniform(5, 20, 2)
t0s = periods * np.random.rand(2)
t = np.arange(0, 80, 0.02)
yerr = 5e-4
Then, define the parameters. In this simple model, we’ll just fit for
the limb darkening parameters of the star, and the period, phase, impact
parameter, and radius ratio of the planets (note: this is already 10
parameters and running MCMC to convergence using
emcee would probably take at least an
hour). For the limb darkening, we’ll use a quadratic law as
parameterized by Kipping (2013)
and for the joint radius ratio and impact parameter distribution we’ll
use the parameterization from Espinoza
(2018). Both of these
reparameterizations are implemented in exoplanet as custom PyMC3
distributions (exoplanet.distributions.QuadLimbDark and
exoplanet.distributions.RadiusImpact respectively).
import pymc3 as pm
with pm.Model() as model:
# The baseline flux
mean = pm.Normal("mean", mu=0.0, sd=1.0)
# The time of a reference transit for each planet
t0 = pm.Normal("t0", mu=t0s, sd=1.0, shape=2)
# The log period; also tracking the period itself
logP = pm.Normal("logP", mu=np.log(periods), sd=0.1, shape=2)
period = pm.Deterministic("period", pm.math.exp(logP))
# The Kipping (2013) parameterization for quadratic limb darkening paramters
u = xo.distributions.QuadLimbDark("u", testval=np.array([0.3, 0.2]))
# The Espinoza (2018) parameterization for the joint radius ratio and
# impact parameter distribution
r, b = xo.distributions.get_joint_radius_impact(
min_radius=0.01, max_radius=0.1,
testval_r=np.array([0.04, 0.06]),
testval_b=np.random.rand(2)
)
# This shouldn't make a huge difference, but I like to put a uniform
# prior on the *log* of the radius ratio instead of the value. This
# can be implemented by adding a custom "potential" (log probability).
pm.Potential("r_prior", -pm.math.log(r))
# Set up a Keplerian orbit for the planets
orbit = xo.orbits.KeplerianOrbit(period=period, t0=t0, b=b)
# Compute the model light curve using starry
light_curves = xo.StarryLightCurve(u).get_light_curve(
orbit=orbit, r=r, t=t)
light_curve = pm.math.sum(light_curves, axis=-1) + mean
# Here we track the value of the model light curve for plotting
# purposes
pm.Deterministic("light_curves", light_curves)
# In this line, we simulate the dataset that we will fit
y = xo.eval_in_model(light_curve)
y += yerr * np.random.randn(len(y))
# The likelihood function assuming known Gaussian uncertainty
pm.Normal("obs", mu=light_curve, sd=yerr, observed=y)
# Fit for the maximum a posteriori parameters given the simuated
# dataset
map_soln = pm.find_MAP(start=model.test_point)
logp = 24,744, ||grad|| = 55,906: 100%|██████████| 28/28 [00:00<00:00, 594.41it/s]
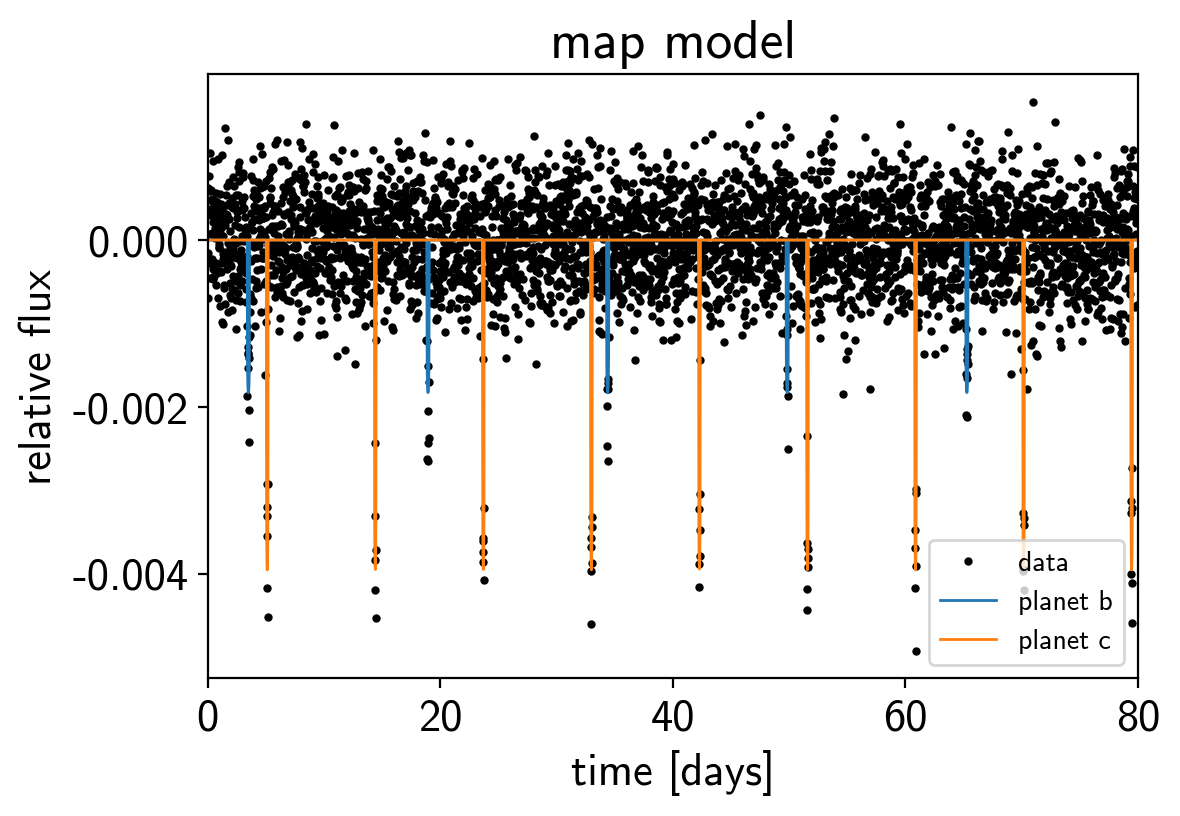
Now we can plot the simulated data and the maximum a posteriori model to make sure that our initialization looks ok.
plt.plot(t, y, ".k", ms=4, label="data")
for i, l in enumerate("bc"):
plt.plot(t, map_soln["light_curves"][:, i], lw=1,
label="planet {0}".format(l))
plt.xlim(t.min(), t.max())
plt.ylabel("relative flux")
plt.xlabel("time [days]")
plt.legend(fontsize=10)
plt.title("map model");
Sampling¶
Now, let’s sample from the posterior defined by this model. As usual,
there are strong covariances between some of the parameters so we’ll use
the exoplanet.PyMC3Sampler to sample.
np.random.seed(42)
sampler = xo.PyMC3Sampler(window=100, finish=200)
with model:
burnin = sampler.tune(tune=2500, start=map_soln, step_kwargs=dict(target_accept=0.9))
trace = sampler.sample(draws=3000)
Only 2 samples in chain.
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [rb, u, logP, t0, mean]
Sampling 2 chains: 100%|██████████| 154/154 [00:18<00:00, 8.42draws/s]
The chain reached the maximum tree depth. Increase max_treedepth, increase target_accept or reparameterize.
The chain reached the maximum tree depth. Increase max_treedepth, increase target_accept or reparameterize.
Only 2 samples in chain.
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [rb, u, logP, t0, mean]
Sampling 2 chains: 100%|██████████| 204/204 [00:22<00:00, 3.35draws/s]
The chain reached the maximum tree depth. Increase max_treedepth, increase target_accept or reparameterize.
The chain reached the maximum tree depth. Increase max_treedepth, increase target_accept or reparameterize.
Only 2 samples in chain.
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [rb, u, logP, t0, mean]
Sampling 2 chains: 100%|██████████| 404/404 [00:39<00:00, 8.14draws/s]
Only 2 samples in chain.
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [rb, u, logP, t0, mean]
Sampling 2 chains: 100%|██████████| 804/804 [00:08<00:00, 97.69draws/s]
Only 2 samples in chain.
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [rb, u, logP, t0, mean]
Sampling 2 chains: 100%|██████████| 3454/3454 [00:34<00:00, 100.80draws/s]
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [rb, u, logP, t0, mean]
Sampling 2 chains: 100%|██████████| 6400/6400 [00:56<00:00, 114.24draws/s]
After sampling, it’s important that we assess convergence. We can do
that using the pymc3.summary function:
pm.summary(trace, varnames=["period", "t0", "r", "b", "u", "mean"])
| mean | sd | mc_error | hpd_2.5 | hpd_97.5 | n_eff | Rhat | |
|---|---|---|---|---|---|---|---|
| period__0 | 15.447790 | 0.001695 | 3.486476e-05 | 15.444912 | 15.451406 | 2533.378312 | 0.999949 |
| period__1 | 9.292520 | 0.000283 | 3.887756e-06 | 9.291975 | 9.293059 | 5548.728925 | 0.999915 |
| t0__0 | 3.501171 | 0.003577 | 5.558718e-05 | 3.494321 | 3.507672 | 4373.233830 | 1.000007 |
| t0__1 | 5.121344 | 0.001412 | 1.877619e-05 | 5.118433 | 5.123968 | 5799.163491 | 0.999900 |
| r__0 | 0.040313 | 0.001091 | 1.657517e-05 | 0.038135 | 0.042390 | 4366.143554 | 1.000034 |
| r__1 | 0.059967 | 0.000882 | 1.446773e-05 | 0.058180 | 0.061578 | 3760.600443 | 0.999938 |
| b__0 | 0.271202 | 0.101419 | 2.313994e-03 | 0.049401 | 0.444910 | 2037.837779 | 0.999845 |
| b__1 | 0.518464 | 0.025133 | 4.471865e-04 | 0.463794 | 0.561249 | 3129.527466 | 0.999834 |
| u__0 | 0.188186 | 0.142469 | 2.078470e-03 | 0.000015 | 0.467653 | 5899.190731 | 1.000029 |
| u__1 | 0.256806 | 0.255075 | 4.093560e-03 | -0.236499 | 0.738364 | 3712.296023 | 0.999841 |
| mean | 0.000004 | 0.000008 | 8.231005e-08 | -0.000011 | 0.000020 | 7493.185005 | 0.999859 |
That looks pretty good! Fitting this without exoplanet would have taken a lot more patience.
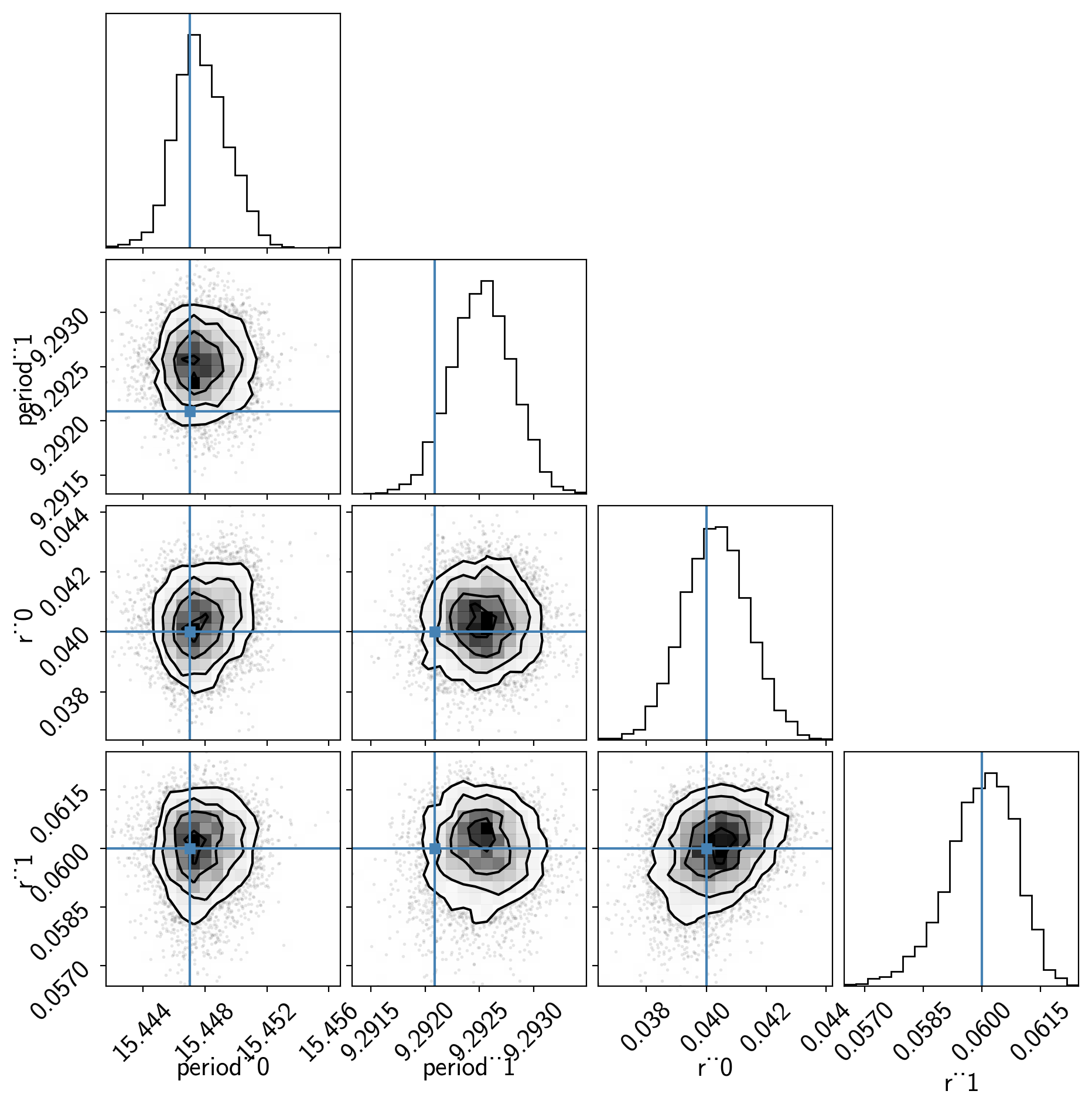
Now we can also look at the corner plot of some of that parameters of interest:
import corner
samples = pm.trace_to_dataframe(trace, varnames=["period", "r"])
truth = np.concatenate(xo.eval_in_model([period, r], model.test_point, model=model))
corner.corner(samples, truths=truth);
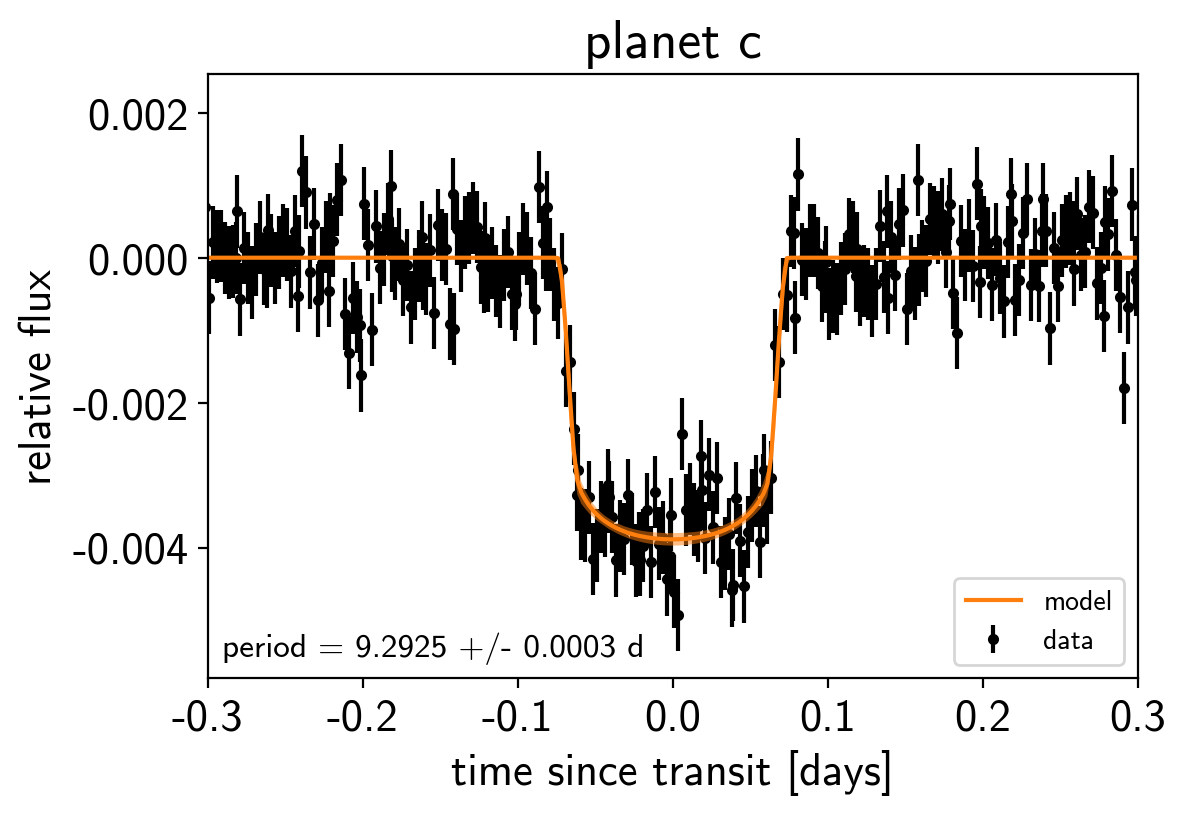
Phase plots¶
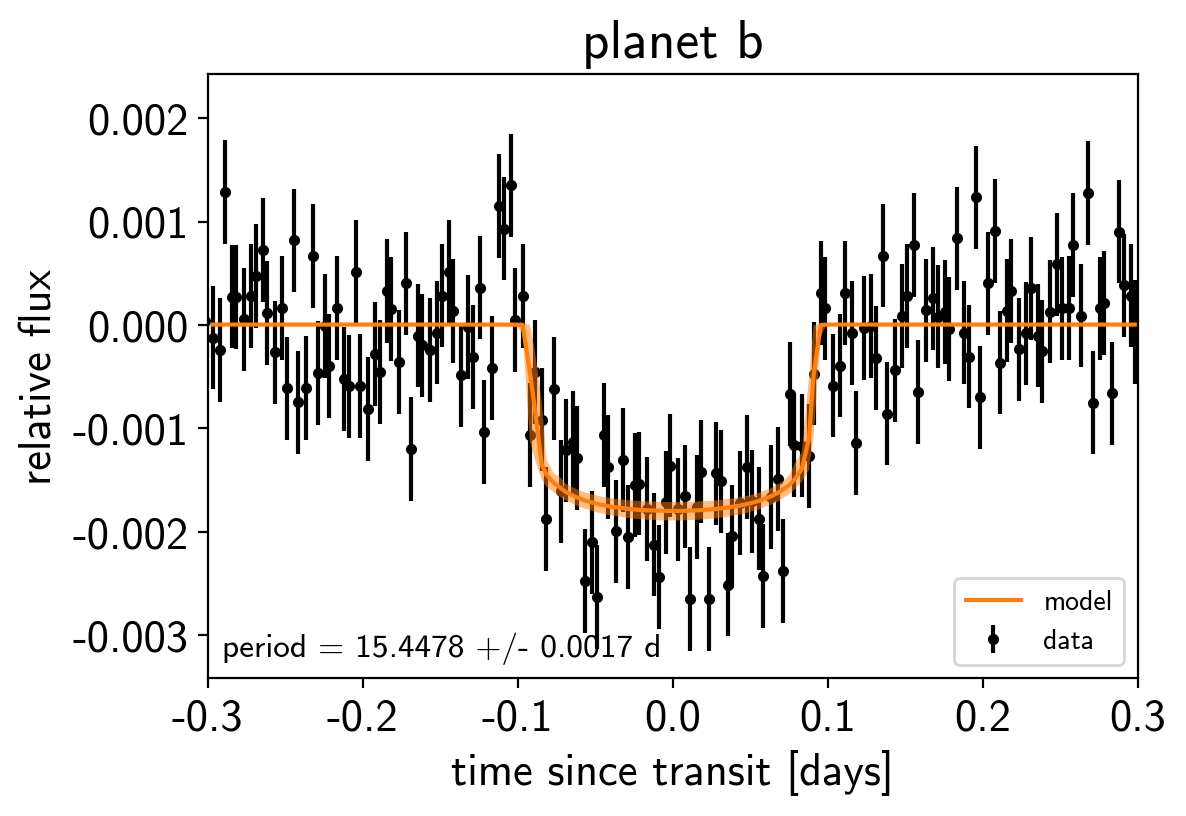
Like in the radial velocity tutorial (Radial velocity fitting), we can make plots of the model predictions for each planet.
for n, letter in enumerate("bc"):
plt.figure()
# Get the posterior median orbital parameters
p = np.median(trace["period"][:, n])
t0 = np.median(trace["t0"][:, n])
# Compute the median of posterior estimate of the contribution from
# the other planet. Then we can remove this from the data to plot
# just the planet we care about.
other = np.median(trace["light_curves"][:, :, (n + 1) % 2], axis=0)
# Plot the folded data
x_fold = (t - t0 + 0.5*p) % p - 0.5*p
plt.errorbar(x_fold, y - other, yerr=yerr, fmt=".k", label="data",
zorder=-1000)
# Plot the folded model
inds = np.argsort(x_fold)
inds = inds[np.abs(x_fold)[inds] < 0.3]
pred = trace["light_curves"][:, inds, n] + trace["mean"][:, None]
pred = np.percentile(pred, [16, 50, 84], axis=0)
plt.plot(x_fold[inds], pred[1], color="C1", label="model")
art = plt.fill_between(x_fold[inds], pred[0], pred[2], color="C1", alpha=0.5,
zorder=1000)
art.set_edgecolor("none")
# Annotate the plot with the planet's period
txt = "period = {0:.4f} +/- {1:.4f} d".format(
np.mean(trace["period"][:, n]), np.std(trace["period"][:, n]))
plt.annotate(txt, (0, 0), xycoords="axes fraction",
xytext=(5, 5), textcoords="offset points",
ha="left", va="bottom", fontsize=12)
plt.legend(fontsize=10, loc=4)
plt.xlim(-0.5*p, 0.5*p)
plt.xlabel("time since transit [days]")
plt.ylabel("relative flux")
plt.title("planet {0}".format(letter));
plt.xlim(-0.3, 0.3)
Citations¶
As described in the Citing exoplanet & its dependencies tutorial, we can use
exoplanet.citations.get_citations_for_model() to construct an
acknowledgement and BibTeX listing that includes the relevant citations
for this model. This is especially important here because we have used
quite a few model components that should be cited.
with model:
txt, bib = xo.citations.get_citations_for_model()
print(txt)
This research made use of textsf{exoplanet} (Foreman-Mackey et al.in prep.) and its dependencies citep{exoplanet:astropy13,exoplanet:as
tropy18,exoplanet:espinoza18,exoplanet:kipping13,exoplanet:luger18,exo
planet:pymc3,exoplanet:theano}.
print("\n".join(bib.splitlines()[:10]) + "\n...")
@article{exoplanet:pymc3,
title={Probabilistic programming in Python using PyMC3},
author={Salvatier, John and Wiecki, Thomas V and Fonnesbeck, Christopher},
journal={PeerJ Computer Science},
volume={2},
pages={e55},
year={2016},
publisher={PeerJ Inc.}
}
...