Note
This tutorial was generated from an IPython notebook that can be downloaded here.
Interpolation with PyMC3¶
A 1D example¶
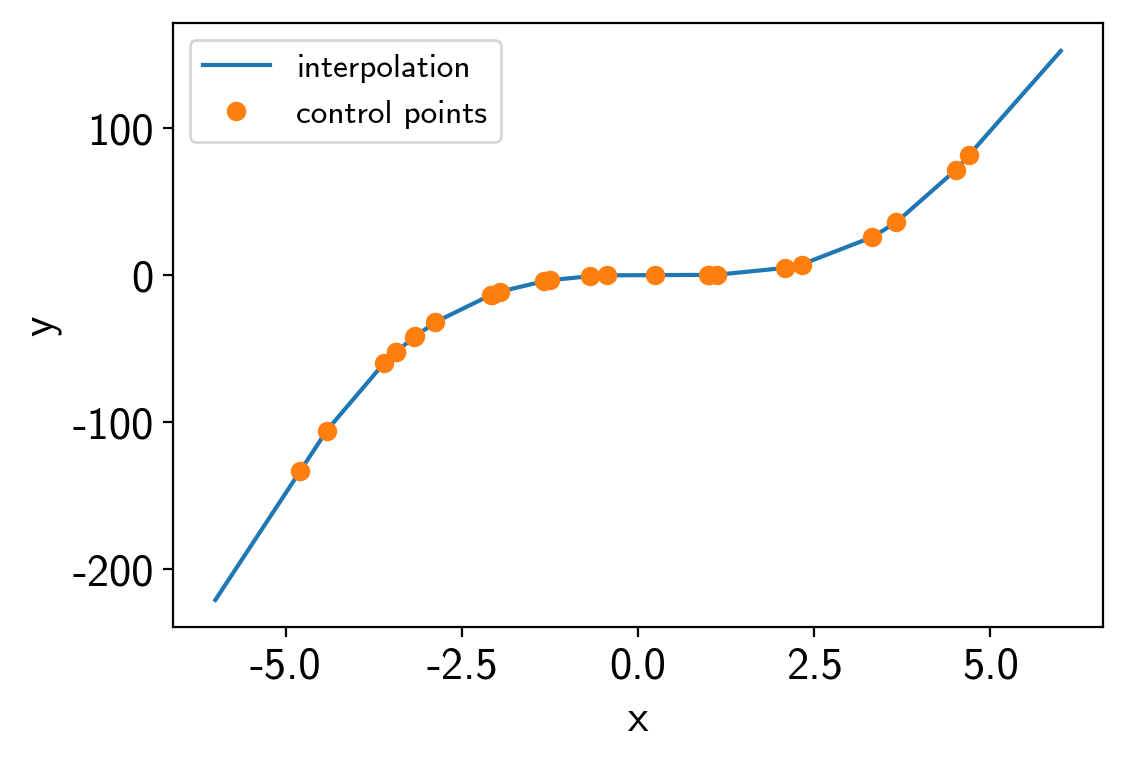
To start, we’ll do a simple 1D example where we have a model evaluated at control points and we interpolate between them to estimate the model value.
import numpy as np
import matplotlib.pyplot as plt
import exoplanet as xo
np.random.seed(42)
x = np.sort(np.random.uniform(-5, 5, 25))
points = [x]
values = x**3-x**2
interpolator = xo.interp.RegularGridInterpolator(points, values[:, None])
t = np.linspace(-6, 6, 5000)
plt.plot(t, interpolator.evaluate(t[:, None]).eval(), label="interpolation")
plt.plot(x, values, "o", label="control points")
plt.xlabel("x")
plt.ylabel("y")
plt.legend(fontsize=12);
Here’s how we build the PyMC3 model:
import pymc3 as pm
truth = 45.0
data_sd = 8.0
data_mu = truth + data_sd * np.random.randn()
with pm.Model() as model:
# The value passed into the interpolator must have the shape
# (ntest, ndim), but in our case that is (1, 1)
xval = pm.Uniform("x", lower=-8, upper=8, shape=(1, 1))
# Evaluate the interpolated model and extract the scalar value
# we want
mod = pm.Deterministic("y", interpolator.evaluate(xval)[0, 0])
# The usual likelihood
pm.Normal("obs", mu=mod, sd=data_sd, observed=data_mu)
# Sampling!
trace = pm.sample(draws=1000, tune=2000, step_kwargs=dict(target_accept=0.9))
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [x]
Sampling 2 chains: 100%|██████████| 6000/6000 [00:02<00:00, 2937.41draws/s]
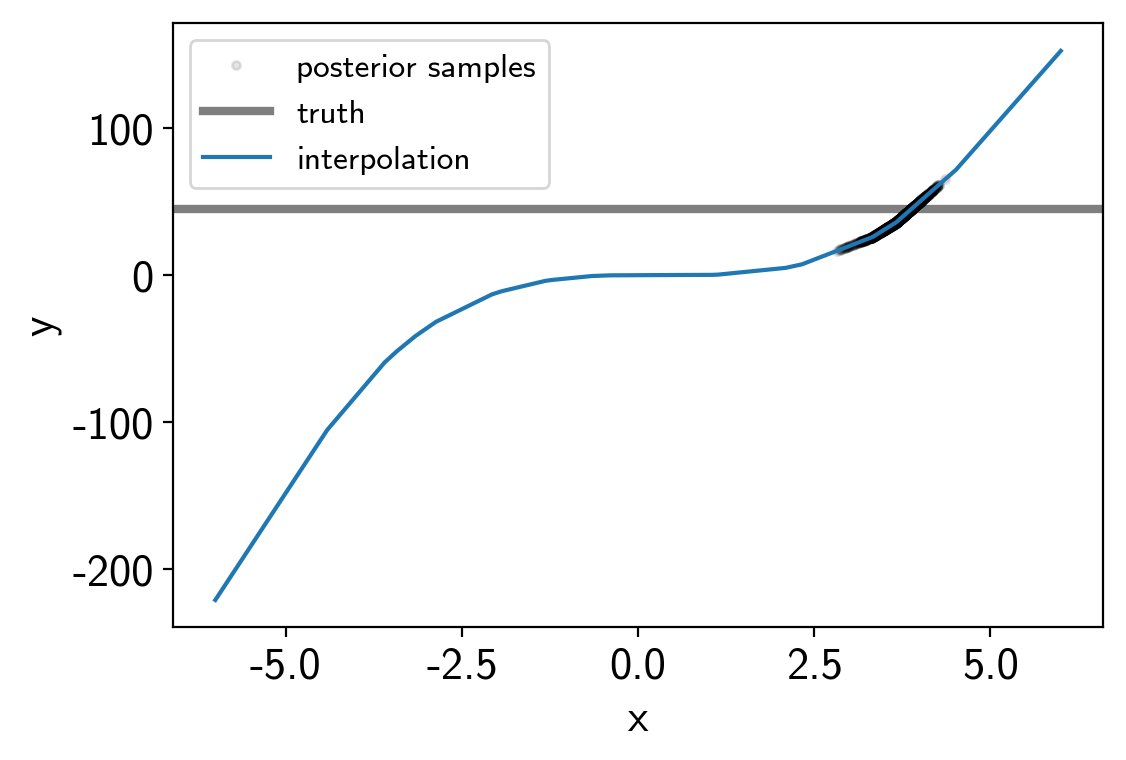
And here are the results:
t = np.linspace(-6, 6, 5000)
plt.plot(trace["x"][:, 0, 0], trace["y"], ".k", alpha=0.1, label="posterior samples")
plt.axhline(truth, color="k", lw=3, alpha=0.5, label="truth")
plt.plot(t, interpolator.evaluate(t[:, None]).eval(), label="interpolation")
plt.xlabel("x")
plt.ylabel("y")
plt.legend(fontsize=12);
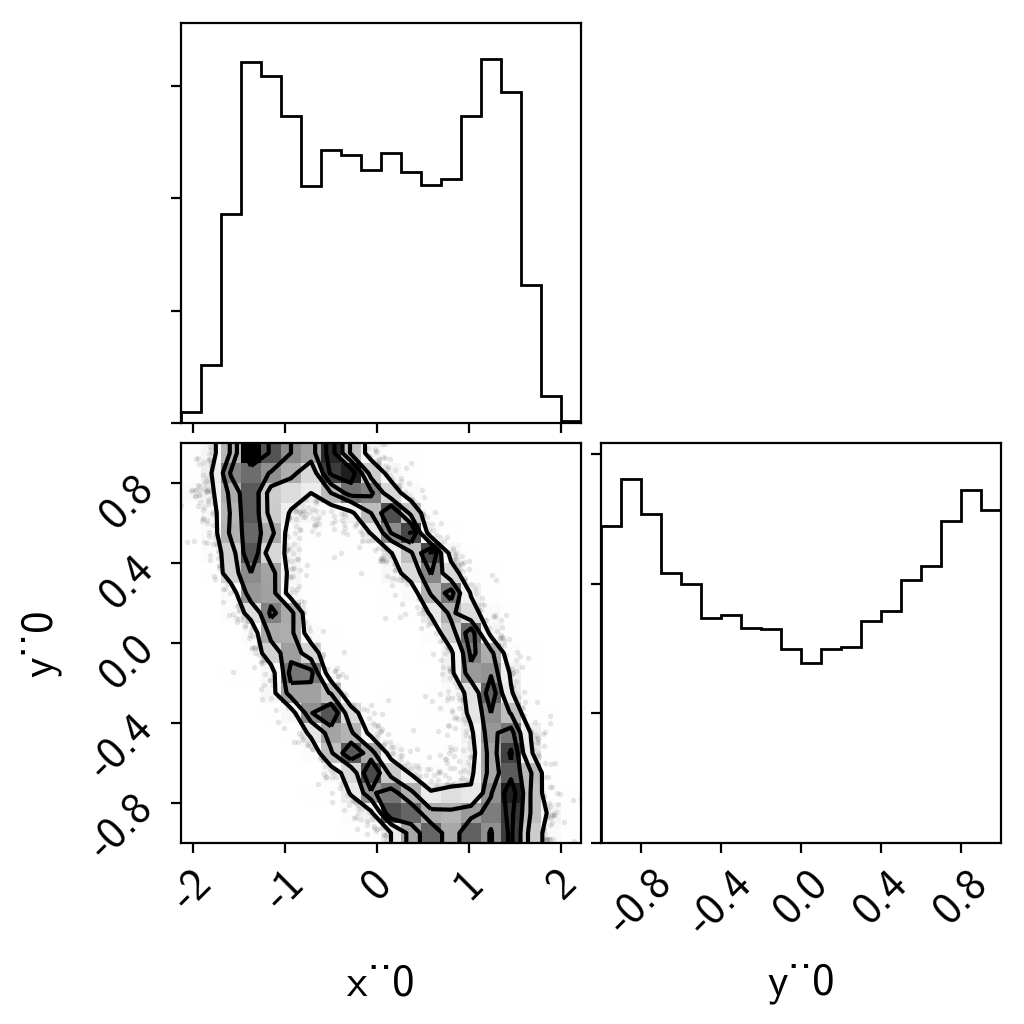
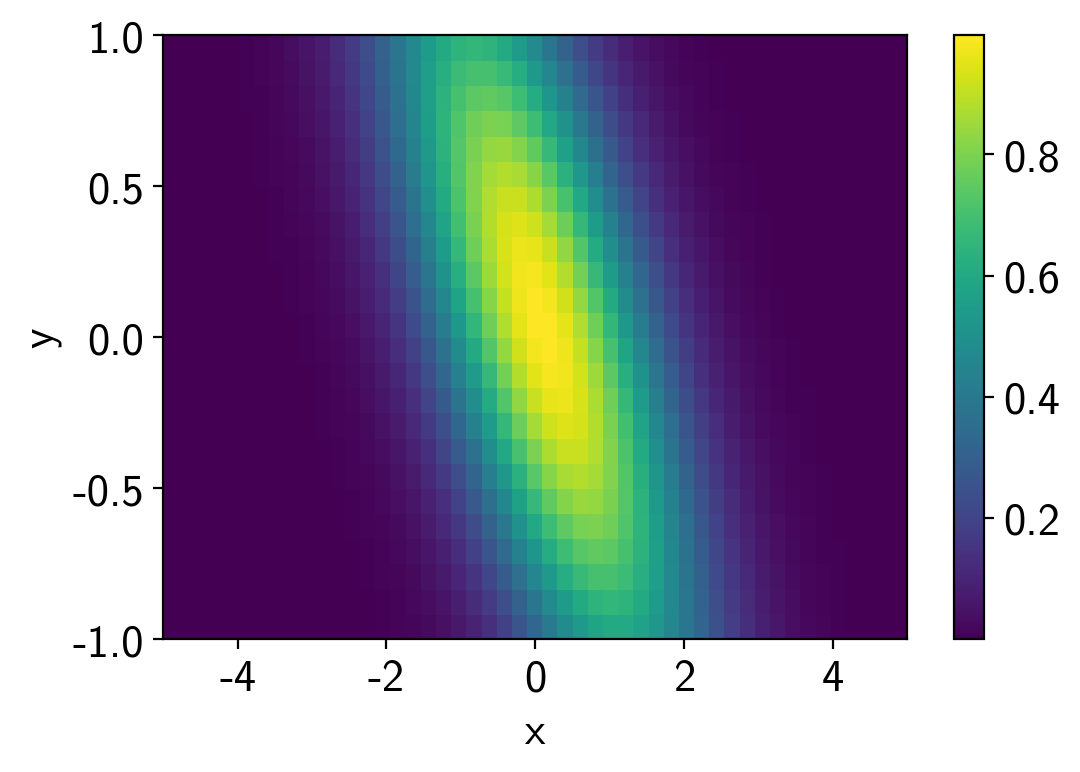
A 2D example¶
In this case, we’ll interpolate a 2D function. This one is a hard one because the posterior is a ring, but it demonstrates the principle.
points = [
np.linspace(-5, 5, 50),
np.linspace(-1, 1, 25),
]
values = np.exp(-0.5*(points[0]**2)[:, None] - 0.5*(points[1]**2 / 0.5)[None, :] - points[0][:, None]*points[1][None, :])
interpolator = xo.interp.RegularGridInterpolator(points, values[:, :, None], nout=1)
plt.pcolor(points[0], points[1], values.T)
plt.colorbar()
plt.xlabel("x")
plt.ylabel("y");
Set things up and sample.
import theano.tensor as tt
data_mu = 0.6
data_sd = 0.1
with pm.Model() as model:
xval = pm.Uniform("x", lower=-5, upper=5, shape=(1,))
yval = pm.Uniform("y", lower=-1, upper=1, shape=(1,))
xtest = tt.stack([xval, yval], axis=-1)
mod = interpolator.evaluate(xtest)
# The usual likelihood
pm.Normal("obs", mu=mod, sd=data_sd, observed=data_mu)
# Sampling!
trace = pm.sample(draws=4000, tune=4000, step_kwargs=dict(target_accept=0.9))
Auto-assigning NUTS sampler...
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (2 chains in 2 jobs)
NUTS: [y, x]
Sampling 2 chains: 100%|██████████| 16000/16000 [00:12<00:00, 1241.36draws/s]
The number of effective samples is smaller than 10% for some parameters.
And here are the results:
import corner
samples = pm.trace_to_dataframe(trace)
corner.corner(samples);